| 1 |
|
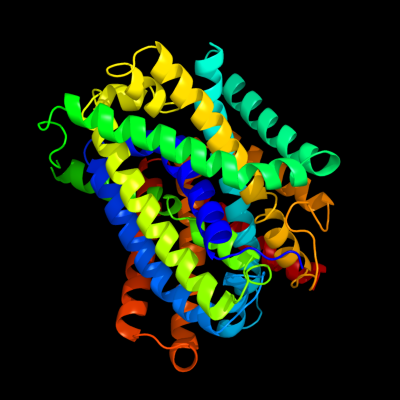
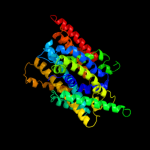
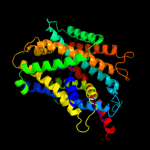
PDB 3gia chain A
Region: 10 - 429
Aligned: 419
Modelled: 419
Confidence: 100.0%
Identity: 31%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 2 |
|
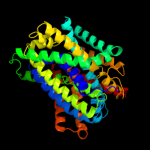
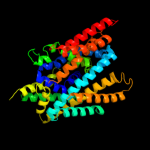
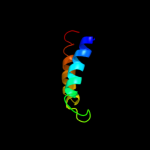
PDB 3lrc chain C
Region: 10 - 429
Aligned: 394
Modelled: 394
Confidence: 100.0%
Identity: 18%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 3 |
|
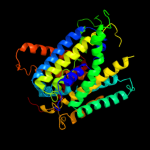
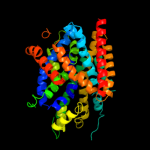
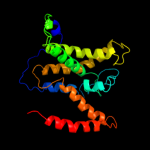
PDB 2jln chain A
Region: 2 - 429
Aligned: 420
Modelled: 428
Confidence: 100.0%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 4 |
|
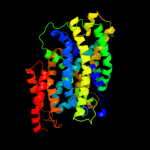
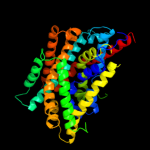
PDB 3dh4 chain A
Region: 11 - 429
Aligned: 413
Modelled: 419
Confidence: 99.2%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 5 |
|
PDB 2xq2 chain A
Region: 11 - 429
Aligned: 410
Modelled: 410
Confidence: 99.2%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 6 |
|
PDB 2a65 chain A domain 1
Region: 14 - 428
Aligned: 410
Modelled: 410
Confidence: 97.6%
Identity: 15%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 7 |
|
PDB 2w8a chain C
Region: 6 - 380
Aligned: 370
Modelled: 375
Confidence: 96.0%
Identity: 11%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 8 |
|
PDB 3hfx chain A
Region: 8 - 380
Aligned: 369
Modelled: 373
Confidence: 87.6%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 9 |
|
PDB 2iub chain A domain 2
Region: 381 - 428
Aligned: 44
Modelled: 48
Confidence: 15.5%
Identity: 11%
Fold: Transmembrane helix hairpin
Superfamily: Magnesium transport protein CorA, transmembrane region
Family: Magnesium transport protein CorA, transmembrane region
Phyre2
| 10 |
|
PDB 2wwb chain A
Region: 210 - 428
Aligned: 213
Modelled: 219
Confidence: 9.5%
Identity: 11%
PDB header:ribosome
Chain: A: PDB Molecule:protein transport protein sec61 subunit alpha isoform 1;
PDBTitle: cryo-em structure of the mammalian sec61 complex bound to the2 actively translating wheat germ 80s ribosome
Phyre2
| 11 |
|
PDB 1ujl chain A
Region: 3 - 24
Aligned: 22
Modelled: 22
Confidence: 9.1%
Identity: 23%
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily h
PDBTitle: solution structure of the herg k+ channel s5-p2 extracellular linker
Phyre2