| 1 |
|
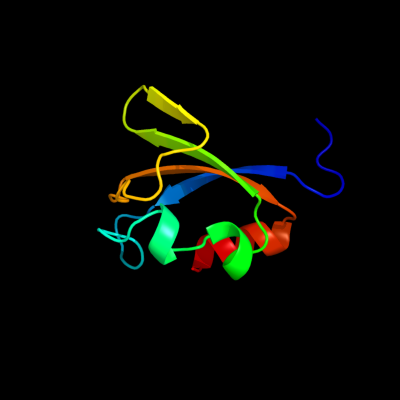
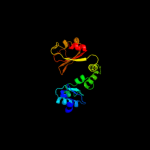
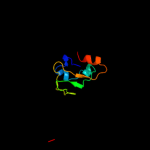
PDB 2fy8 chain A domain 2
Region: 319 - 395
Aligned: 77
Modelled: 77
Confidence: 99.4%
Identity: 25%
Fold: TrkA C-terminal domain-like
Superfamily: TrkA C-terminal domain-like
Family: TrkA C-terminal domain-like
Phyre2
| 2 |
|
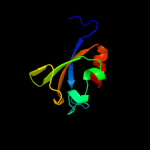
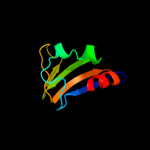
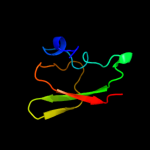
PDB 1vct chain A domain 2
Region: 320 - 394
Aligned: 75
Modelled: 75
Confidence: 99.4%
Identity: 21%
Fold: TrkA C-terminal domain-like
Superfamily: TrkA C-terminal domain-like
Family: TrkA C-terminal domain-like
Phyre2
| 3 |
|
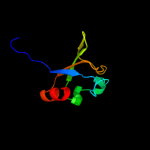
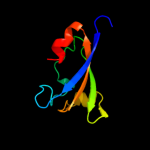
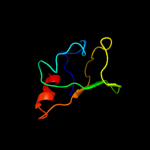
PDB 2bkn chain A
Region: 320 - 394
Aligned: 75
Modelled: 75
Confidence: 99.3%
Identity: 21%
PDB header:membrane protein
Chain: A: PDB Molecule:hypothetical protein ph0236;
PDBTitle: structure analysis of unknown function protein
Phyre2
| 4 |
|
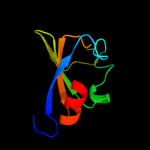
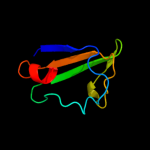
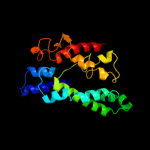
PDB 1lnq chain C
Region: 216 - 394
Aligned: 169
Modelled: 179
Confidence: 98.9%
Identity: 17%
PDB header:metal transport
Chain: C: PDB Molecule:potassium channel related protein;
PDBTitle: crystal structure of mthk at 3.3 a
Phyre2
| 5 |
|
PDB 2fy8 chain A
Region: 320 - 394
Aligned: 75
Modelled: 75
Confidence: 98.9%
Identity: 25%
PDB header:transport protein
Chain: A: PDB Molecule:calcium-gated potassium channel mthk;
PDBTitle: crystal structure of mthk rck domain in its ligand-free gating-ring2 form
Phyre2
| 6 |
|
PDB 3jxo chain B
Region: 321 - 394
Aligned: 71
Modelled: 74
Confidence: 98.8%
Identity: 17%
PDB header:transport protein
Chain: B: PDB Molecule:trka-n domain protein;
PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
Phyre2
| 7 |
|
PDB 3l4b chain G
Region: 328 - 394
Aligned: 63
Modelled: 67
Confidence: 97.4%
Identity: 16%
PDB header:transport protein
Chain: G: PDB Molecule:trka k+ channel protien tm1088b;
PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
Phyre2
| 8 |
|
PDB 3u6n chain C
Region: 331 - 397
Aligned: 67
Modelled: 67
Confidence: 97.0%
Identity: 12%
PDB header:transport protein
Chain: C: PDB Molecule:high-conductance ca2+-activated k+ channel protein;
PDBTitle: open structure of the bk channel gating ring
Phyre2
| 9 |
|
PDB 3mt5 chain A
Region: 331 - 385
Aligned: 55
Modelled: 55
Confidence: 95.5%
Identity: 13%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:potassium large conductance calcium-activated channel,
PDBTitle: crystal structure of the human bk gating apparatus
Phyre2
| 10 |
|
PDB 3naf chain A
Region: 331 - 395
Aligned: 65
Modelled: 65
Confidence: 72.3%
Identity: 14%
PDB header:ion transport
Chain: A: PDB Molecule:calcium-activated potassium channel subunit alpha-1;
PDBTitle: structure of the intracellular gating ring from the human high-2 conductance ca2+ gated k+ channel (bk channel)
Phyre2
| 11 |
|
PDB 1l7v chain A
Region: 7 - 199
Aligned: 174
Modelled: 193
Confidence: 20.0%
Identity: 13%
Fold: ABC transporter involved in vitamin B12 uptake, BtuC
Superfamily: ABC transporter involved in vitamin B12 uptake, BtuC
Family: ABC transporter involved in vitamin B12 uptake, BtuC
Phyre2
| 12 |
|
PDB 2nq2 chain A
Region: 56 - 200
Aligned: 127
Modelled: 145
Confidence: 13.1%
Identity: 16%
PDB header:metal transport
Chain: A: PDB Molecule:hypothetical abc transporter permease protein
PDBTitle: an inward-facing conformation of a putative metal-chelate2 type abc transporter.
Phyre2
| 13 |
|
PDB 2c2x chain B
Region: 328 - 371
Aligned: 44
Modelled: 44
Confidence: 12.9%
Identity: 11%
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase-
PDBTitle: three dimensional structure of bifunctional2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase3 from mycobacterium tuberculosis
Phyre2