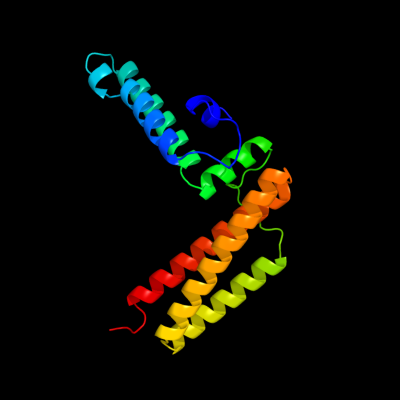
| 1 | c1fpoA_
|
|
 |
100.0 |
99 |
PDB header:chaperone
Chain: A: PDB Molecule:chaperone protein hscb;
PDBTitle: hsc20 (hscb), a j-type co-chaperone from e. coli
|
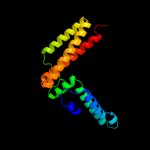
| 2 | c3bvoA_
|
|
 |
100.0 |
31 |
PDB header:chaperone
Chain: A: PDB Molecule:co-chaperone protein hscb, mitochondrial precursor;
PDBTitle: crystal structure of human co-chaperone protein hscb
|
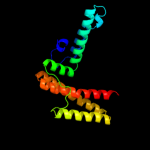
| 3 | c3hhoA_
|
|
 |
100.0 |
48 |
PDB header:chaperone
Chain: A: PDB Molecule:co-chaperone protein hscb homolog;
PDBTitle: chaperone hscb from vibrio cholerae
|
| 4 | c3uo2A_
|
|
 |
99.9 |
28 |
PDB header:chaperone
Chain: A: PDB Molecule:j-type co-chaperone jac1, mitochondrial;
PDBTitle: jac1 co-chaperone from saccharomyces cerevisiae
|
| 5 | d1fpoa1
|
|
 |
99.9 |
100 |
Fold:Long alpha-hairpin
Superfamily:Chaperone J-domain
Family:Chaperone J-domain |
| 6 | c3apqB_
|
|
 |
99.8 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dnaj homolog subfamily c member 10;
PDBTitle: crystal structure of j-trx1 fragment of erdj5
|
| 7 | c2l6lA_
|
|
 |
99.8 |
28 |
PDB header:chaperone
Chain: A: PDB Molecule:dnaj homolog subfamily c member 24;
PDBTitle: solution structure of human j-protein co-chaperone, dph4
|
| 8 | c2ctqA_
|
|
 |
99.8 |
25 |
PDB header:chaperone
Chain: A: PDB Molecule:dnaj homolog subfamily c member 12;
PDBTitle: solution structure of j-domain from human dnaj subfamily c2 menber 12
|
| 9 | c2qsaA_
|
|
 |
99.8 |
23 |
PDB header:chaperone
Chain: A: PDB Molecule:dnaj homolog dnj-2;
PDBTitle: crystal structure of j-domain of dnaj homolog dnj-2 precursor from2 c.elegans.
|
| 10 | c2cugA_
|
|
 |
99.7 |
22 |
PDB header:chaperone
Chain: A: PDB Molecule:mkiaa0962 protein;
PDBTitle: solution structure of the j domain of the pseudo dnaj2 protein, mouse hypothetical mkiaa0962
|
| 11 | d1wjza_
|
|
 |
99.7 |
27 |
Fold:Long alpha-hairpin
Superfamily:Chaperone J-domain
Family:Chaperone J-domain |
| 12 | d1gh6a_
|
|
 |
99.7 |
16 |
Fold:Long alpha-hairpin
Superfamily:Chaperone J-domain
Family:Chaperone J-domain |
| 13 | c2yuaA_
|
|
 |
99.7 |
24 |
PDB header:chaperone
Chain: A: PDB Molecule:williams-beuren syndrome chromosome region 18
PDBTitle: solution structure of the dnaj domain from human williams-2 beuren syndrome chromosome region 18 protein
|
| 14 | c2ctwA_
|
|
 |
99.7 |
25 |
PDB header:chaperone
Chain: A: PDB Molecule:dnaj homolog subfamily c member 5;
PDBTitle: solution structure of j-domain from mouse dnaj subfamily c2 menber 5
|
| 15 | c2kqxA_
|
|
 |
99.7 |
27 |
PDB header:chaperone binding protein
Chain: A: PDB Molecule:curved dna-binding protein;
PDBTitle: nmr structure of the j-domain (residues 2-72) in the2 escherichia coli cbpa
|
| 16 | d1hdja_
|
|
 |
99.7 |
22 |
Fold:Long alpha-hairpin
Superfamily:Chaperone J-domain
Family:Chaperone J-domain |
| 17 | c2dmxA_
|
|
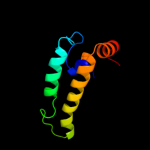 |
99.7 |
18 |
PDB header:chaperone
Chain: A: PDB Molecule:dnaj homolog subfamily b member 8;
PDBTitle: solution structure of the j domain of dnaj homolog2 subfamily b member 8
|
| 18 | c2lgwA_
|
|
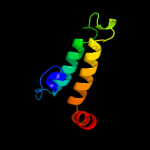 |
99.7 |
18 |
PDB header:chaperone
Chain: A: PDB Molecule:dnaj homolog subfamily b member 2;
PDBTitle: solution structure of the j domain of hsj1a
|
| 19 | d1xbla_
|
|
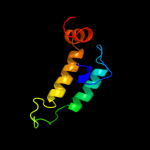 |
99.7 |
25 |
Fold:Long alpha-hairpin
Superfamily:Chaperone J-domain
Family:Chaperone J-domain |
| 20 | c2dn9A_
|
|
 |
99.7 |
21 |
PDB header:apoptosis, chaperone
Chain: A: PDB Molecule:dnaj homolog subfamily a member 3;
PDBTitle: solution structure of j-domain from the dnaj homolog, human2 tid1 protein
|
| 21 | c2ctrA_ |
|
not modelled |
99.7 |
30 |
PDB header:chaperone
Chain: A: PDB Molecule:dnaj homolog subfamily b member 9;
PDBTitle: solution structure of j-domain from human dnaj subfamily b2 menber 9
|
| 22 | c2ctpA_ |
|
not modelled |
99.7 |
25 |
PDB header:chaperone
Chain: A: PDB Molecule:dnaj homolog subfamily b member 12;
PDBTitle: solution structure of j-domain from human dnaj subfamily b2 menber 12
|
| 23 | c2o37A_ |
|
not modelled |
99.7 |
24 |
PDB header:chaperone
Chain: A: PDB Molecule:protein sis1;
PDBTitle: j-domain of sis1 protein, hsp40 co-chaperone from2 saccharomyces cerevisiae.
|
| 24 | c2ochA_ |
|
not modelled |
99.7 |
23 |
PDB header:chaperone
Chain: A: PDB Molecule:hypothetical protein dnj-12;
PDBTitle: j-domain of dnj-12 from caenorhabditis elegans
|
| 25 | d1fafa_ |
|
not modelled |
99.7 |
22 |
Fold:Long alpha-hairpin
Superfamily:Chaperone J-domain
Family:Chaperone J-domain |
| 26 | c1bq0A_ |
|
not modelled |
99.6 |
25 |
PDB header:chaperone
Chain: A: PDB Molecule:dnaj;
PDBTitle: j-domain (residues 1-77) of the escherichia coli n-terminal2 fragment (residues 1-104) of the molecular chaperone dnaj,3 nmr, 20 structures
|
| 27 | c2ys8A_ |
|
not modelled |
99.6 |
16 |
PDB header:protein binding
Chain: A: PDB Molecule:rab-related gtp-binding protein rabj;
PDBTitle: solution structure of the dnaj-like domain from human ras-2 associated protein rap1
|
| 28 | c2pf4E_ |
|
not modelled |
99.6 |
14 |
PDB header:hydrolase regulator/viral protein
Chain: E: PDB Molecule:small t antigen;
PDBTitle: crystal structure of the full-length simian virus 40 small t antigen2 complexed with the protein phosphatase 2a aalpha subunit
|
| 29 | d1iura_ |
|
not modelled |
99.6 |
13 |
Fold:Long alpha-hairpin
Superfamily:Chaperone J-domain
Family:Chaperone J-domain |
| 30 | d1nz6a_ |
|
not modelled |
99.4 |
16 |
Fold:Long alpha-hairpin
Superfamily:Chaperone J-domain
Family:Chaperone J-domain |
| 31 | d1n4ca_ |
|
not modelled |
99.4 |
17 |
Fold:Long alpha-hairpin
Superfamily:Chaperone J-domain
Family:Chaperone J-domain |
| 32 | d1fpoa2 |
|
not modelled |
99.4 |
99 |
Fold:Open three-helical up-and-down bundle
Superfamily:HSC20 (HSCB), C-terminal oligomerisation domain
Family:HSC20 (HSCB), C-terminal oligomerisation domain |
| 33 | c2guzO_ |
|
not modelled |
99.3 |
21 |
PDB header:chaperone, protein transport
Chain: O: PDB Molecule:mitochondrial import inner membrane translocase
PDBTitle: structure of the tim14-tim16 complex of the mitochondrial2 protein import motor
|
| 34 | c3ag7A_ |
|
not modelled |
99.2 |
18 |
PDB header:plant protein
Chain: A: PDB Molecule:putative uncharacterized protein f9e10.5;
PDBTitle: an auxilin-like j-domain containing protein, jac1 j-domain
|
| 35 | c2y4tA_ |
|
not modelled |
99.1 |
29 |
PDB header:chaperone
Chain: A: PDB Molecule:dnaj homolog subfamily c member 3;
PDBTitle: crystal structure of the human co-chaperone p58(ipk)
|
| 36 | c2guzD_ |
|
not modelled |
99.1 |
17 |
PDB header:chaperone, protein transport
Chain: D: PDB Molecule:mitochondrial import inner membrane translocase
PDBTitle: structure of the tim14-tim16 complex of the mitochondrial2 protein import motor
|
| 37 | c3apoA_ |
|
not modelled |
98.8 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dnaj homolog subfamily c member 10;
PDBTitle: crystal structure of full-length erdj5
|
| 38 | d1ug2a_ |
|
not modelled |
63.3 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Myb/SANT domain |
| 39 | d1fc2c_ |
|
not modelled |
30.8 |
16 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Bacterial immunoglobulin/albumin-binding domains
Family:Immunoglobulin-binding protein A modules |
| 40 | d1deeg_ |
|
not modelled |
24.2 |
13 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Bacterial immunoglobulin/albumin-binding domains
Family:Immunoglobulin-binding protein A modules |
| 41 | d1lp1b_ |
|
not modelled |
23.7 |
18 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Bacterial immunoglobulin/albumin-binding domains
Family:Immunoglobulin-binding protein A modules |
| 42 | d2jwda1 |
|
not modelled |
20.7 |
18 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Bacterial immunoglobulin/albumin-binding domains
Family:Immunoglobulin-binding protein A modules |
| 43 | d1ni7a_ |
|
not modelled |
20.4 |
13 |
Fold:SufE/NifU
Superfamily:SufE/NifU
Family:SufE-like |
| 44 | d2d00a1 |
|
not modelled |
14.6 |
19 |
Fold:AtpF-like
Superfamily:AtpF-like
Family:AtpF-like |
| 45 | c3epvB_ |
|
not modelled |
14.1 |
19 |
PDB header:metal binding protein
Chain: B: PDB Molecule:nickel and cobalt resistance protein cnrr;
PDBTitle: x-ray structure of the metal-sensor cnrx in both the apo- and copper-2 bound forms
|
| 46 | c2zfdB_ |
|
not modelled |
12.2 |
18 |
PDB header:signaling protein/transferase
Chain: B: PDB Molecule:putative uncharacterized protein t20l15_90;
PDBTitle: the crystal structure of plant specific calcium binding protein atcbl22 in complex with the regulatory domain of atcipk14
|
| 47 | c2ehbD_ |
|
not modelled |
11.4 |
24 |
PDB header:signalling protein/transferase
Chain: D: PDB Molecule:cbl-interacting serine/threonine-protein kinase 24;
PDBTitle: the structure of the c-terminal domain of the protein kinase atsos22 bound to the calcium sensor atsos3
|
| 48 | d2etva1 |
|
not modelled |
10.9 |
12 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:TM0189-like |
| 49 | c2vrgA_ |
|
not modelled |
10.1 |
16 |
PDB header:transport
Chain: A: PDB Molecule:multiple coagulation factor deficiency protein 2;
PDBTitle: structure of human mcfd2
|
| 50 | d1pmia_ |
|
not modelled |
9.8 |
44 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Type I phosphomannose isomerase |
| 51 | c3pshA_ |
|
not modelled |
9.7 |
16 |
PDB header:metal transport
Chain: A: PDB Molecule:protein hi_1472;
PDBTitle: classification of a haemophilus influenzae abc transporter hi1470/712 through its cognate molybdate periplasmic binding protein mola (mola3 bound to molybdate)
|
| 52 | c3h1yA_ |
|
not modelled |
8.8 |
36 |
PDB header:isomerase
Chain: A: PDB Molecule:mannose-6-phosphate isomerase;
PDBTitle: crystal structure of mannose 6-phosphate isomerase from2 salmonella typhimurium bound to substrate (f6p)and metal3 atom (zn)
|
| 53 | c2l53B_ |
|
not modelled |
8.7 |
14 |
PDB header:ca-binding protein/proton transport
Chain: B: PDB Molecule:voltage-gated sodium channel type v alpha isoform b
PDBTitle: solution nmr structure of apo-calmodulin in complex with the iq motif2 of human cardiac sodium channel nav1.5
|
| 54 | d2py6a1 |
|
not modelled |
8.6 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:FkbM-like |
| 55 | c2j8kA_ |
|
not modelled |
8.5 |
19 |
PDB header:toxin
Chain: A: PDB Molecule:np275-np276;
PDBTitle: structure of the fusion of np275 and np276, pentapeptide2 repeat proteins from nostoc punctiforme
|
| 56 | d2gtad1 |
|
not modelled |
8.3 |
15 |
Fold:all-alpha NTP pyrophosphatases
Superfamily:all-alpha NTP pyrophosphatases
Family:MazG-like |
| 57 | d1uf2a_ |
|
not modelled |
8.2 |
33 |
Fold:Reovirus inner layer core protein p3
Superfamily:Reovirus inner layer core protein p3
Family:Phytoreovirus core |
| 58 | c1uf2A_ |
|
not modelled |
8.2 |
33 |
PDB header:virus
Chain: A: PDB Molecule:core protein p3;
PDBTitle: the atomic structure of rice dwarf virus (rdv)
|
| 59 | c2kxwB_ |
|
not modelled |
8.1 |
0 |
PDB header:calcium-binding protein/metal transport
Chain: B: PDB Molecule:sodium channel protein type 2 subunit alpha;
PDBTitle: structure of the c-domain fragment of apo calmodulin bound to the iq2 motif of nav1.2
|
| 60 | d1edla_ |
|
not modelled |
8.0 |
14 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Bacterial immunoglobulin/albumin-binding domains
Family:Immunoglobulin-binding protein A modules |
| 61 | d1c20a_ |
|
not modelled |
7.8 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:ARID-like
Family:ARID domain |
| 62 | c2rq5A_ |
|
not modelled |
7.6 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:protein jumonji;
PDBTitle: solution structure of the at-rich interaction domain (arid)2 of jumonji/jarid2
|
| 63 | d1mxaa1 |
|
not modelled |
7.1 |
100 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 64 | d2p02a1 |
|
not modelled |
6.7 |
100 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 65 | c2eqyA_ |
|
not modelled |
6.6 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:jumonji, at rich interactive domain 1b;
PDBTitle: solution structure of the arid domain of jarid1b protein
|
| 66 | d2dara2 |
|
not modelled |
6.5 |
40 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 67 | d3saka_ |
|
not modelled |
6.4 |
17 |
Fold:p53 tetramerization domain
Superfamily:p53 tetramerization domain
Family:p53 tetramerization domain |
| 68 | c2ov6A_ |
|
not modelled |
6.4 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:v-type atp synthase subunit f;
PDBTitle: the nmr structure of subunit f of the methanogenic a1ao atp synthase2 and its interaction with the nucleotide-binding subunit b
|
| 69 | c3fn2A_ |
|
not modelled |
6.2 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:putative sensor histidine kinase domain;
PDBTitle: crystal structure of a putative sensor histidine kinase domain from2 clostridium symbiosum atcc 14940
|
| 70 | d1boda_ |
|
not modelled |
6.2 |
18 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calbindin D9K |
| 71 | c1b0nB_ |
|
not modelled |
6.0 |
0 |
PDB header:transcription regulator
Chain: B: PDB Molecule:protein (sini protein);
PDBTitle: sinr protein/sini protein complex
|
| 72 | d1b0nb_ |
|
not modelled |
6.0 |
0 |
Fold:Dimerisation interlock
Superfamily:SinR repressor dimerisation domain-like
Family:SinR repressor dimerisation domain-like |
| 73 | d3fapb_ |
|
not modelled |
6.0 |
12 |
Fold:Four-helical up-and-down bundle
Superfamily:FKBP12-rapamycin-binding domain of FKBP-rapamycin-associated protein (FRAP)
Family:FKBP12-rapamycin-binding domain of FKBP-rapamycin-associated protein (FRAP) |
| 74 | d1bh9b_ |
|
not modelled |
5.8 |
13 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:TBP-associated factors, TAFs |
| 75 | d1p77a2 |
|
not modelled |
5.7 |
15 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Shikimate dehydrogenase-like |
| 76 | d1ig6a_ |
|
not modelled |
5.4 |
4 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:ARID-like
Family:ARID domain |
| 77 | d1qwra_ |
|
not modelled |
5.3 |
25 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Type I phosphomannose isomerase |
| 78 | d1cf7a_ |
|
not modelled |
5.3 |
26 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Cell cycle transcription factor e2f-dp |
| 79 | d2oeea1 |
|
not modelled |
5.2 |
11 |
Fold:YheA-like
Superfamily:YheA/YmcA-like
Family:YheA-like |
| 80 | c2jrzA_ |
|
not modelled |
5.2 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:histone demethylase jarid1c;
PDBTitle: solution structure of the bright/arid domain from the human2 jarid1c protein.
|