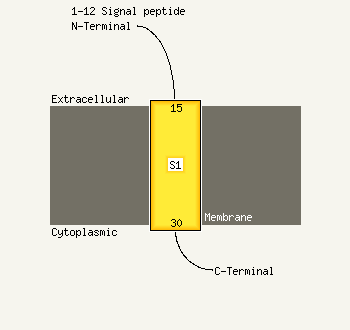
1 c2vxgB_
34.1
13
PDB header: gene regulationChain: B: PDB Molecule: cg6181-pa, isoform a;PDBTitle: crystal structure of the conserved c-terminal region of ge-2 1
2 c1v06A_
21.3
33
PDB header: dna-binding proteinChain: A: PDB Molecule: hmg box-containing protein 1;PDBTitle: axh domain of the transcription factor hbp1 from m.musculus
3 c2px2B_
12.3
38
PDB header: transferaseChain: B: PDB Molecule: genome polyprotein [contains: capsid protein cPDBTitle: crystal structure of the murray valley encephalitis virus2 ns5 2'-o methyltransferase domain in complex with sah3 (monoclinic form 1)
4 c3gczA_
11.3
38
PDB header: transferaseChain: A: PDB Molecule: polyprotein;PDBTitle: yokose virus methyltransferase in complex with adomet
5 c2yu1A_
10.3
28
PDB header: oxidoreductaseChain: A: PDB Molecule: jmjc domain-containing histone demethylation protein 1a;PDBTitle: crystal structure of hjhdm1a complexed with a-ketoglutarate
6 c2ds2A_
10.3
19
PDB header: plant proteinChain: A: PDB Molecule: sweet protein mabinlin-2 chain a;PDBTitle: crystal structure of mabinlin ii
7 c2kixD_
10.2
21
PDB header: transport proteinChain: D: PDB Molecule: bm2 protein;PDBTitle: channel domain of bm2 protein from influenza b virus
8 c3n9mC_
10.2
22
PDB header: oxidoreductaseChain: C: PDB Molecule: putative uncharacterized protein;PDBTitle: cekdm7a from c.elegans, alone
9 d1yela1
10.0
13
Fold: DNA-binding pseudobarrel domainSuperfamily: DNA-binding pseudobarrel domainFamily: B3 DNA binding domain10 c3evaA_
9.6
25
PDB header: transferaseChain: A: PDB Molecule: rna-directed rna polymerase ns5;PDBTitle: crystal structure of yellow fever virus methyltransferase complexed2 with s-adenosyl-l-homocysteine
11 d1i94l_
9.4
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like12 c1wazA_
8.8
40
PDB header: transport proteinChain: A: PDB Molecule: merf;PDBTitle: nmr structure determination of the bacterial mercury2 transporter, merf, in micelles
13 c3lkzB_
8.8
20
PDB header: viral proteinChain: B: PDB Molecule: non-structural protein 5;PDBTitle: structural and functional analyses of a conserved hydrophobic pocket2 of flavivirus methyltransferase
14 d2p41a1
8.5
40
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: mRNA cap methylase15 c2jo1A_
7.5
25
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
16 c2jp3A_
7.3
24
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
17 d1s3ga2
7.0
21
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain18 d1zina2
6.7
21
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain19 c2h3oA_
6.6
40
PDB header: membrane proteinChain: A: PDB Molecule: merf;PDBTitle: structure of merft, a membrane protein with two trans-2 membrane helices
20 d1p3ja2
6.5
29
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain21 d2uubl1
not modelled
6.1
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like22 d1t1ra3
not modelled
6.1
75
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain23 c3pu3A_
not modelled
5.7
33
PDB header: protein bindingChain: A: PDB Molecule: phd finger protein 2;PDBTitle: phf2 jumonji domain-nog complex
24 d1xvha1
not modelled
5.7
22
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Bacterial immunoglobulin/albumin-binding domainsFamily: GA module, an albumin-binding domain25 d2qall1
not modelled
5.6
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like26 c3pesA_
not modelled
5.5
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein gp49;PDBTitle: crystal structure of uncharacterized protein from pseudomonas phage2 yua
27 c1ujlA_
not modelled
5.5
50
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily hPDBTitle: solution structure of the herg k+ channel s5-p2 extracellular linker
28 c2p3qA_
not modelled
5.3
50
PDB header: viral protein,transferaseChain: A: PDB Molecule: type ii methyltransferase;PDBTitle: crystal structure of dengue methyltransferase in complex with gpppg2 and s-adenosyl-l-homocysteine
29 d1akya2
not modelled
5.2
29
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain30 c3kv5D_
not modelled
5.2
33
PDB header: h3k4me3 binding protein, transferaseChain: D: PDB Molecule: jmjc domain-containing histone demethylationPDBTitle: structure of kiaa1718, human jumonji demethylase, in complex2 with n-oxalylglycine
31 c3alrA_
not modelled
5.1
33
PDB header: metal binding proteinChain: A: PDB Molecule: nanos protein;PDBTitle: crystal structure of nanos