| 1 |
|
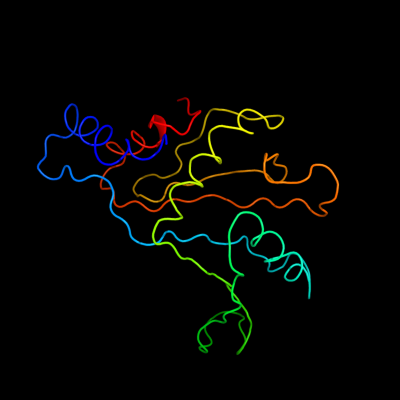
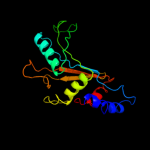
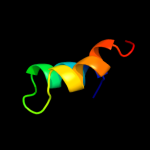
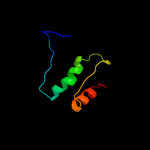
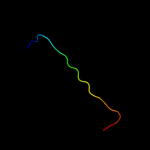
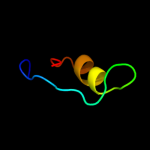
PDB 1iq4 chain A
Region: 1 - 179
Aligned: 179
Modelled: 179
Confidence: 100.0%
Identity: 61%
Fold: RL5-like
Superfamily: RL5-like
Family: Ribosomal protein L5
Phyre2
| 2 |
|
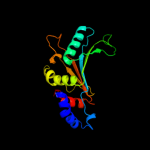
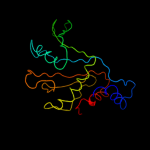
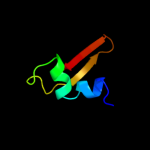
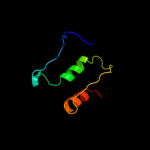
PDB 1mji chain A
Region: 3 - 179
Aligned: 177
Modelled: 177
Confidence: 100.0%
Identity: 56%
Fold: RL5-like
Superfamily: RL5-like
Family: Ribosomal protein L5
Phyre2
| 3 |
|
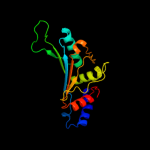
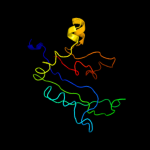
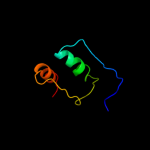
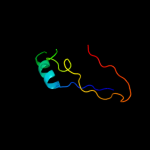
PDB 2zjr chain D domain 1
Region: 3 - 179
Aligned: 177
Modelled: 177
Confidence: 100.0%
Identity: 60%
Fold: RL5-like
Superfamily: RL5-like
Family: Ribosomal protein L5
Phyre2
| 4 |
|
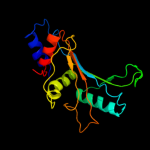
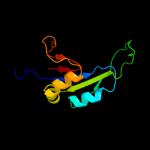
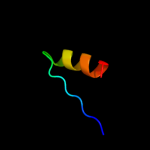
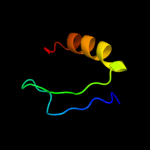
PDB 3bbo chain H
Region: 3 - 177
Aligned: 175
Modelled: 175
Confidence: 100.0%
Identity: 49%
PDB header:ribosome
Chain: H: PDB Molecule:ribosomal protein l5;
PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
Phyre2
| 5 |
|
PDB 2gyc chain D domain 1
Region: 3 - 179
Aligned: 177
Modelled: 177
Confidence: 100.0%
Identity: 100%
Fold: RL5-like
Superfamily: RL5-like
Family: Ribosomal protein L5
Phyre2
| 6 |
|
PDB 4a1c chain D
Region: 21 - 167
Aligned: 137
Modelled: 147
Confidence: 100.0%
Identity: 26%
PDB header:ribosome
Chain: D: PDB Molecule:60s ribosomal protein l11;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
Phyre2
| 7 |
|
PDB 1s1i chain J
Region: 24 - 159
Aligned: 126
Modelled: 136
Confidence: 100.0%
Identity: 34%
PDB header:ribosome
Chain: J: PDB Molecule:60s ribosomal protein l11;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
Phyre2
| 8 |
|
PDB 1vqo chain D domain 1
Region: 24 - 138
Aligned: 99
Modelled: 107
Confidence: 100.0%
Identity: 42%
Fold: RL5-like
Superfamily: RL5-like
Family: Ribosomal protein L5
Phyre2
| 9 |
|
PDB 2rf4 chain B
Region: 91 - 119
Aligned: 29
Modelled: 29
Confidence: 38.8%
Identity: 38%
PDB header:transferase
Chain: B: PDB Molecule:dna-directed rna polymerase i subunit rpa4;
PDBTitle: crystal structure of the rna polymerase i subcomplex a14/43
Phyre2
| 10 |
|
PDB 3d37 chain A domain 1
Region: 93 - 131
Aligned: 30
Modelled: 39
Confidence: 17.9%
Identity: 27%
Fold: Phage tail proteins
Superfamily: Phage tail proteins
Family: Baseplate protein-like
Phyre2
| 11 |
|
PDB 2jvf chain A
Region: 153 - 173
Aligned: 21
Modelled: 21
Confidence: 17.5%
Identity: 33%
PDB header:de novo protein
Chain: A: PDB Molecule:de novo protein m7;
PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
Phyre2
| 12 |
|
PDB 3hxx chain A
Region: 116 - 176
Aligned: 60
Modelled: 61
Confidence: 17.5%
Identity: 22%
PDB header:ligase
Chain: A: PDB Molecule:alanyl-trna synthetase;
PDBTitle: crystal structure of catalytic fragment of e. coli alars in complex2 with amppcp
Phyre2
| 13 |
|
PDB 1qys chain A
Region: 153 - 173
Aligned: 21
Modelled: 21
Confidence: 17.1%
Identity: 24%
PDB header:de novo protein
Chain: A: PDB Molecule:top7;
PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
Phyre2
| 14 |
|
PDB 1yfs chain B
Region: 116 - 176
Aligned: 59
Modelled: 61
Confidence: 12.8%
Identity: 25%
PDB header:ligase
Chain: B: PDB Molecule:alanyl-trna synthetase;
PDBTitle: the crystal structure of alanyl-trna synthetase in complex2 with l-alanine
Phyre2
| 15 |
|
PDB 1riq chain A domain 2
Region: 116 - 176
Aligned: 59
Modelled: 61
Confidence: 10.0%
Identity: 24%
Fold: Class II aaRS and biotin synthetases
Superfamily: Class II aaRS and biotin synthetases
Family: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain
Phyre2
| 16 |
|
PDB 2x6v chain B
Region: 86 - 133
Aligned: 44
Modelled: 48
Confidence: 9.2%
Identity: 23%
PDB header:transcription/dna
Chain: B: PDB Molecule:t-box transcription factor tbx5;
PDBTitle: crystal structure of human tbx5 in the dna-bound and dna-2 free form
Phyre2
| 17 |
|
PDB 1vjq chain B
Region: 140 - 177
Aligned: 38
Modelled: 38
Confidence: 8.6%
Identity: 24%
PDB header:structural genomics, de novo protein
Chain: B: PDB Molecule:designed protein;
PDBTitle: designed protein based on backbone conformation of2 procarboxypeptidase-a (1aye) with sidechains chosen for maximal3 predicted stability.
Phyre2
| 18 |
|
PDB 3c25 chain A
Region: 109 - 128
Aligned: 20
Modelled: 20
Confidence: 6.4%
Identity: 30%
PDB header:hydrolase/dna
Chain: A: PDB Molecule:noti restriction endonuclease;
PDBTitle: crystal structure of noti restriction endonuclease bound to cognate2 dna
Phyre2
| 19 |
|
PDB 2xzn chain H
Region: 147 - 169
Aligned: 23
Modelled: 23
Confidence: 6.2%
Identity: 26%
PDB header:ribosome
Chain: H: PDB Molecule:ribosomal protein s8 containing protein;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
Phyre2
| 20 |
|
PDB 1xtz chain A
Region: 151 - 178
Aligned: 25
Modelled: 28
Confidence: 5.6%
Identity: 12%
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase;
PDBTitle: crystal structure of the s. cerevisiae d-ribose-5-phosphate isomerase:2 comparison with the archeal and bacterial enzymes
Phyre2
| 21 |
|
| 22 |
|