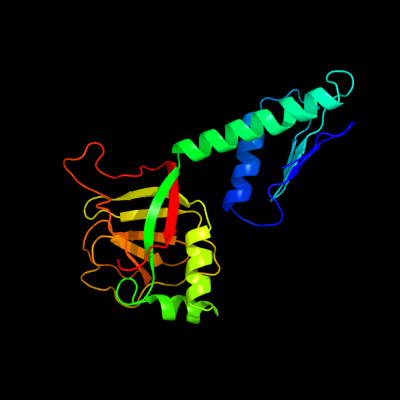
| 1 | c2phcB_
|
|
 |
100.0 |
39 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein ph0987;
PDBTitle: crystal structure of conserved uncharacterized protein ph0987 from2 pyrococcus horikoshii
|
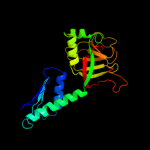
| 2 | c3oepA_
|
|
 |
100.0 |
38 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative uncharacterized protein ttha0988;
PDBTitle: crystal structure of ttha0988 in space group p43212
|
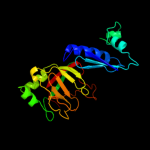
| 3 | c3mmlD_
|
|
 |
100.0 |
37 |
PDB header:hydrolase
Chain: D: PDB Molecule:allophanate hydrolase subunit 1;
PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
|
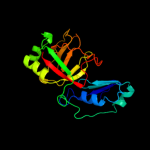
| 4 | d2phcb1
|
|
 |
100.0 |
54 |
Fold:Cyclophilin-like
Superfamily:Cyclophilin-like
Family:PH0987 C-terminal domain-like |
| 5 | c2zp2B_
|
|
 |
100.0 |
57 |
PDB header:transferase inhibitor
Chain: B: PDB Molecule:kinase a inhibitor;
PDBTitle: c-terminal domain of kipi from bacillus subtilis
|
| 6 | c2kwaA_
|
|
 |
99.5 |
17 |
PDB header:transferase inhibitor
Chain: A: PDB Molecule:kinase a inhibitor;
PDBTitle: 1h, 13c and 15n backbone and side chain resonance assignments of the2 n-terminal domain of the histidine kinase inhibitor kipi from3 bacillus subtilis
|
| 7 | d2phcb2
|
|
 |
99.5 |
17 |
Fold:DCoH-like
Superfamily:PH0987 N-terminal domain-like
Family:PH0987 N-terminal domain-like |
| 8 | c2qf7A_
|
|
 |
94.4 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase protein;
PDBTitle: crystal structure of a complete multifunctional pyruvate carboxylase2 from rhizobium etli
|
| 9 | c2j89A_
|
|
 |
88.3 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine sulfoxide reductase a;
PDBTitle: functional and structural aspects of poplar cytosolic and2 plastidial type a methionine sulfoxide reductases
|
| 10 | c1nwaA_
|
|
 |
86.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:peptide methionine sulfoxide reductase msra;
PDBTitle: structure of mycobacterium tuberculosis methionine2 sulfoxide reductase a in complex with protein-bound3 methionine
|
| 11 | d1nwaa_
|
|
 |
86.9 |
17 |
Fold:Ferredoxin-like
Superfamily:Peptide methionine sulfoxide reductase
Family:Peptide methionine sulfoxide reductase |
| 12 | c3mmlE_
|
|
 |
83.3 |
24 |
PDB header:hydrolase
Chain: E: PDB Molecule:allophanate hydrolase subunit 2;
PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
|
| 13 | c3e0mB_
|
|
 |
78.8 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peptide methionine sulfoxide reductase msra/msrb
PDBTitle: crystal structure of fusion protein of msra and msrb
|
| 14 | c2ropA_
|
|
 |
76.1 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:copper-transporting atpase 2;
PDBTitle: solution structure of domains 3 and 4 of human atp7b
|
| 15 | c3bqhA_
|
|
 |
73.3 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:peptide methionine sulfoxide reductase msra/msrb;
PDBTitle: structure of the central domain (msra) of neisseria meningitidis pilb2 (oxidized form)
|
| 16 | d1afia_
|
|
 |
72.7 |
9 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 17 | c2ew9A_
|
|
 |
72.7 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:copper-transporting atpase 2;
PDBTitle: solution structure of apowln5-6
|
| 18 | c2rmlA_
|
|
 |
71.5 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:copper-transporting p-type atpase copa;
PDBTitle: solution structure of the n-terminal soluble domains of2 bacillus subtilis copa
|
| 19 | d1p6ta2
|
|
 |
69.6 |
22 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 20 | d1s6ua_
|
|
 |
69.2 |
7 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 21 | d1q8la_ |
|
not modelled |
68.1 |
4 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 22 | c3dxsX_ |
|
not modelled |
63.6 |
13 |
PDB header:hydrolase
Chain: X: PDB Molecule:copper-transporting atpase ran1;
PDBTitle: crystal structure of a copper binding domain from hma7, a p-2 type atpase
|
| 23 | d1kvja_ |
|
not modelled |
63.5 |
16 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 24 | c2kkhA_ |
|
not modelled |
62.3 |
13 |
PDB header:metal transport
Chain: A: PDB Molecule:putative heavy metal transporter;
PDBTitle: structure of the zinc binding domain of the atpase hma4
|
| 25 | d2qifa1 |
|
not modelled |
61.6 |
7 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 26 | d1whza_ |
|
not modelled |
61.6 |
20 |
Fold:dsRBD-like
Superfamily:YcfA/nrd intein domain
Family:YcfA-like |
| 27 | d1ff3a_ |
|
not modelled |
61.6 |
10 |
Fold:Ferredoxin-like
Superfamily:Peptide methionine sulfoxide reductase
Family:Peptide methionine sulfoxide reductase |
| 28 | c2ldiA_ |
|
not modelled |
61.6 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:zinc-transporting atpase;
PDBTitle: nmr solution structure of ziaan sub mutant
|
| 29 | c2ga7A_ |
|
not modelled |
61.0 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:copper-transporting atpase 1;
PDBTitle: solution structure of the copper(i) form of the third metal-2 binding domain of atp7a protein (menkes disease protein)
|
| 30 | d1ff3c_ |
|
not modelled |
60.6 |
10 |
Fold:Ferredoxin-like
Superfamily:Peptide methionine sulfoxide reductase
Family:Peptide methionine sulfoxide reductase |
| 31 | c1yjrA_ |
|
not modelled |
60.4 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:copper-transporting atpase 1;
PDBTitle: solution structure of the apo form of the sixth soluble2 domain a69p mutant of menkes protein
|
| 32 | c2l3mA_ |
|
not modelled |
60.1 |
7 |
PDB header:metal binding protein
Chain: A: PDB Molecule:copper-ion-binding protein;
PDBTitle: solution structure of the putative copper-ion-binding protein from2 bacillus anthracis str. ames
|
| 33 | c1fvaA_ |
|
not modelled |
59.8 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:peptide methionine sulfoxide reductase;
PDBTitle: crystal structure of bovine methionine sulfoxide reductase
|
| 34 | d1osda_ |
|
not modelled |
59.7 |
7 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 35 | c3pilA_ |
|
not modelled |
58.5 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:peptide methionine sulfoxide reductase;
PDBTitle: crystal structure of mxr1 from saccharomyces cerevisiae in reduced2 form
|
| 36 | c2ofhX_ |
|
not modelled |
57.6 |
16 |
PDB header:hydrolase, membrane protein
Chain: X: PDB Molecule:zinc-transporting atpase;
PDBTitle: solution structure of the n-terminal domain of the zinc(ii) atpase2 ziaa in its apo form
|
| 37 | c1y3kA_ |
|
not modelled |
55.8 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:copper-transporting atpase 1;
PDBTitle: solution structure of the apo form of the fifth domain of2 menkes protein
|
| 38 | d1cpza_ |
|
not modelled |
53.6 |
11 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 39 | c2dy3B_ |
|
not modelled |
51.4 |
25 |
PDB header:isomerase
Chain: B: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of alanine racemase from corynebacterium glutamicum
|
| 40 | d1fvga_ |
|
not modelled |
49.9 |
10 |
Fold:Ferredoxin-like
Superfamily:Peptide methionine sulfoxide reductase
Family:Peptide methionine sulfoxide reductase |
| 41 | c1l0oC_ |
|
not modelled |
46.6 |
35 |
PDB header:protein binding
Chain: C: PDB Molecule:sigma factor;
PDBTitle: crystal structure of the bacillus stearothermophilus anti-2 sigma factor spoiiab with the sporulation sigma factor3 sigmaf
|
| 42 | d1l0oc_ |
|
not modelled |
46.6 |
35 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
| 43 | d2cbpa_ |
|
not modelled |
41.7 |
27 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 44 | d1rp3a1 |
|
not modelled |
39.1 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma3 domain |
| 45 | c3j09A_ |
|
not modelled |
37.9 |
4 |
PDB header:hydrolase, metal transport
Chain: A: PDB Molecule:copper-exporting p-type atpase a;
PDBTitle: high resolution helical reconstruction of the bacterial p-type atpase2 copper transporter copa
|
| 46 | c3gwqB_ |
|
not modelled |
37.8 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:d-serine deaminase;
PDBTitle: crystal structure of a putative d-serine deaminase (bxe_a4060) from2 burkholderia xenovorans lb400 at 2.00 a resolution
|
| 47 | d1mdah_ |
|
not modelled |
37.2 |
25 |
Fold:7-bladed beta-propeller
Superfamily:YVTN repeat-like/Quinoprotein amine dehydrogenase
Family:Methylamine dehydrogenase, H-chain |
| 48 | d3cnxa1 |
|
not modelled |
36.5 |
38 |
Fold:Cystatin-like
Superfamily:NTF2-like
Family:SAV4671-like |
| 49 | c2k6zA_ |
|
not modelled |
34.9 |
36 |
PDB header:metal transport
Chain: A: PDB Molecule:putative uncharacterized protein ttha1943;
PDBTitle: solution structures of copper loaded form pcua (trans2 conformation of the peptide bond involving the nitrogen of3 p14)
|
| 50 | d1pina2 |
|
not modelled |
32.1 |
36 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 51 | c1yw5A_ |
|
not modelled |
31.5 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:peptidyl prolyl cis/trans isomerase;
PDBTitle: peptidyl-prolyl isomerase ess1 from candida albicans
|
| 52 | d1m5ya3 |
|
not modelled |
29.9 |
45 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 53 | d1eq3a_ |
|
not modelled |
29.4 |
36 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 54 | d1mv8a1 |
|
not modelled |
29.1 |
20 |
Fold:6-phosphogluconate dehydrogenase C-terminal domain-like
Superfamily:6-phosphogluconate dehydrogenase C-terminal domain-like
Family:UDP-glucose/GDP-mannose dehydrogenase dimerisation domain |
| 55 | d2aw0a_ |
|
not modelled |
28.9 |
9 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 56 | d1ku2a1 |
|
not modelled |
27.4 |
27 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma3 domain |
| 57 | c1zeqX_ |
|
not modelled |
27.0 |
33 |
PDB header:metal binding protein
Chain: X: PDB Molecule:cation efflux system protein cusf;
PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
|
| 58 | d1p6ta1 |
|
not modelled |
26.7 |
18 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 59 | d1dlja1 |
|
not modelled |
26.5 |
25 |
Fold:6-phosphogluconate dehydrogenase C-terminal domain-like
Superfamily:6-phosphogluconate dehydrogenase C-terminal domain-like
Family:UDP-glucose/GDP-mannose dehydrogenase dimerisation domain |
| 60 | c3h87D_ |
|
not modelled |
26.4 |
26 |
PDB header:toxin/antitoxin
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: rv0301 rv0300 toxin antitoxin complex from mycobacterium tuberculosis
|
| 61 | c3h25A_ |
|
not modelled |
26.2 |
12 |
PDB header:replication/dna
Chain: A: PDB Molecule:replication protein b;
PDBTitle: crystal structure of the catalytic domain of primase repb' in complex2 with initiator dna
|
| 62 | d1z67a1 |
|
not modelled |
25.4 |
13 |
Fold:YidB-like
Superfamily:YidB-like
Family:YidB-like |
| 63 | c2q3eH_ |
|
not modelled |
24.2 |
8 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: structure of human udp-glucose dehydrogenase complexed with nadh and2 udp-glucose
|
| 64 | c2rqsA_ |
|
not modelled |
24.1 |
27 |
PDB header:isomerase
Chain: A: PDB Molecule:parvulin-like peptidyl-prolyl isomerase;
PDBTitle: 3d structure of pin from the psychrophilic archeon cenarcheaum2 symbiosum (cspin)
|
| 65 | c3m9yB_ |
|
not modelled |
23.4 |
23 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from methicillin2 resistant staphylococcus aureus at 1.9 angstrom resolution
|
| 66 | c1f8aB_ |
|
not modelled |
23.0 |
36 |
PDB header:isomerase
Chain: B: PDB Molecule:peptidyl-prolyl cis-trans isomerase nima-
PDBTitle: structural basis for the phosphoserine-proline recognition2 by group iv ww domains
|
| 67 | c2l55A_ |
|
not modelled |
22.7 |
50 |
PDB header:metal binding protein
Chain: A: PDB Molecule:silb,silver efflux protein, mfp component of the three
PDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
|
| 68 | c1zk6A_ |
|
not modelled |
22.5 |
36 |
PDB header:isomerase
Chain: A: PDB Molecule:foldase protein prsa;
PDBTitle: nmr solution structure of b. subtilis prsa ppiase
|
| 69 | c2jzvA_ |
|
not modelled |
22.4 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:foldase protein prsa;
PDBTitle: solution structure of s. aureus prsa-ppiase
|
| 70 | d1wi9a_ |
|
not modelled |
21.6 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:PCI domain (PINT motif) |
| 71 | d1x9la_ |
|
not modelled |
21.5 |
28 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:DR1885-like metal-binding protein
Family:DR1885-like metal-binding protein |
| 72 | d1xnea_ |
|
not modelled |
21.2 |
21 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:ProFAR isomerase associated domain |
| 73 | c1kzlA_ |
|
not modelled |
20.6 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:riboflavin synthase;
PDBTitle: riboflavin synthase from s.pombe bound to2 carboxyethyllumazine
|
| 74 | c2odoC_ |
|
not modelled |
19.9 |
26 |
PDB header:isomerase
Chain: C: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of pseudomonas fluorescens alanine racemase
|
| 75 | d1r2ra_ |
|
not modelled |
19.8 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 76 | d2ggpb1 |
|
not modelled |
19.5 |
20 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 77 | d1m6ja_ |
|
not modelled |
19.4 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 78 | d2pv2a1 |
|
not modelled |
19.2 |
18 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 79 | c3s6dA_ |
|
not modelled |
19.0 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:putative triosephosphate isomerase;
PDBTitle: crystal structure of a putative triosephosphate isomerase from2 coccidioides immitis
|
| 80 | d1n7va_ |
|
not modelled |
18.3 |
40 |
Fold:Adsorption protein p2
Superfamily:Adsorption protein p2
Family:Adsorption protein p2 |
| 81 | d1j6ya_ |
|
not modelled |
18.2 |
27 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 82 | c2k53A_ |
|
not modelled |
18.1 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:a3dk08 protein;
PDBTitle: nmr solution structure of a3dk08 protein from clostridium2 thermocellum: northeast structural genomics consortium3 target cmr9
|
| 83 | c3a35B_ |
|
not modelled |
18.1 |
14 |
PDB header:luminescent protein
Chain: B: PDB Molecule:lumazine protein;
PDBTitle: crystal structure of lump complexed with riboflavin
|
| 84 | d2btma_ |
|
not modelled |
18.1 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 85 | c3gpkA_ |
|
not modelled |
17.5 |
27 |
PDB header:isomerase
Chain: A: PDB Molecule:ppic-type peptidyl-prolyl cis-trans isomerase;
PDBTitle: crystal structure of ppic-type peptidyl-prolyl cis-trans isomerase2 domain at 1.55a resolution.
|
| 86 | c3jt0B_ |
|
not modelled |
16.9 |
21 |
PDB header:structural protein
Chain: B: PDB Molecule:lamin-b1;
PDBTitle: crystal structure of the c-terminal fragment (426-558)2 lamin-b1 from homo sapiens, northeast structural genomics3 consortium target hr5546a
|
| 87 | c2kqaA_ |
|
not modelled |
16.8 |
58 |
PDB header:toxin
Chain: A: PDB Molecule:cerato-platanin;
PDBTitle: the solution structure of the fungal elicitor cerato-platanin
|
| 88 | d2rcda1 |
|
not modelled |
16.8 |
28 |
Fold:Cystatin-like
Superfamily:NTF2-like
Family:BxeB1374-like |
| 89 | d1ndsa2 |
|
not modelled |
16.6 |
19 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 90 | c3kspA_ |
|
not modelled |
16.5 |
27 |
PDB header:unknown function
Chain: A: PDB Molecule:calcium/calmodulin-dependent kinase ii association domain;
PDBTitle: crystal structure of a putative ca/calmodulin-dependent kinase ii2 association domain (exig_1688) from exiguobacterium sibiricum 255-153 at 2.59 a resolution
|
| 91 | c2kt2A_ |
|
not modelled |
16.4 |
7 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mercuric reductase;
PDBTitle: structure of nmera, the n-terminal hma domain of tn501 mercuric2 reductase
|
| 92 | c2pv3B_ |
|
not modelled |
16.4 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:chaperone sura;
PDBTitle: crystallographic structure of sura fragment lacking the second2 peptidyl-prolyl isomerase domain complexed with peptide nftlkfwdifrk
|
| 93 | c3cieC_ |
|
not modelled |
16.4 |
27 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glyceraldehyde-3-phosphate dehydrogenase;
PDBTitle: crystal structure of glyceraldehyde 3-phosphate2 dehydrogenase from cryptosporidium parvum
|
| 94 | d1jnsa_ |
|
not modelled |
16.0 |
45 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 95 | d1o7ia_ |
|
not modelled |
16.0 |
21 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Single strand DNA-binding domain, SSB |
| 96 | c3krsB_ |
|
not modelled |
15.9 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: structure of triosephosphate isomerase from cryptosporidium parvum at2 1.55a resolution
|
| 97 | c2kgjA_ |
|
not modelled |
15.6 |
9 |
PDB header:isomerase
Chain: A: PDB Molecule:peptidyl-prolyl cis-trans isomerase d;
PDBTitle: solution structure of parvulin domain of ppid from e.coli
|
| 98 | c2becA_ |
|
not modelled |
15.6 |
29 |
PDB header:metal binding protein/transport protein
Chain: A: PDB Molecule:calcineurin b homologous protein 2;
PDBTitle: crystal structure of chp2 in complex with its binding2 region in nhe1 and insights into the mechanism of ph3 regulation
|
| 99 | c3th6B_ |
|
not modelled |
15.4 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from rhipicephalus2 (boophilus) microplus.
|