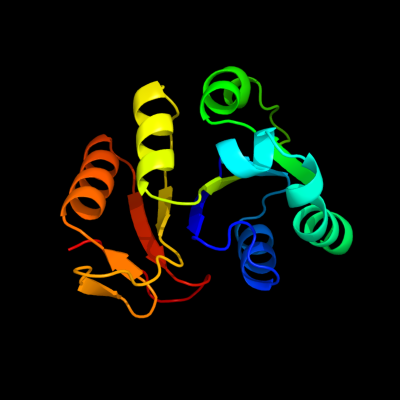
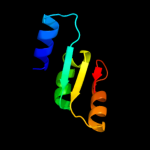
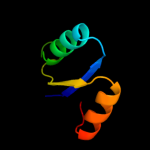
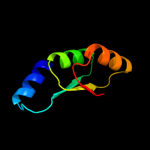
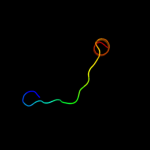
1 c3eyeA_
100.0
100
PDB header: transferaseChain: A: PDB Molecule: pts system n-acetylgalactosamine-specific iibPDBTitle: crystal structure of pts system n-acetylgalactosamine-2 specific iib component 1 from escherichia coli
2 d1nrza_
100.0
30
Fold: PTS IIb componentSuperfamily: PTS IIb componentFamily: PTS IIb component3 d1blea_
100.0
28
Fold: PTS IIb componentSuperfamily: PTS IIb componentFamily: PTS IIb component4 c3lfjB_
100.0
25
PDB header: transferaseChain: B: PDB Molecule: phosphotransferase system, mannose/fructose/n-PDBTitle: crystal structure of manxb from thermoanaerobacter tengcongensis
5 c3p3vB_
100.0
34
PDB header: transferaseChain: B: PDB Molecule: pts system, n-acetylgalactosamine-specific iib component;PDBTitle: crystal structure of a pts dependent n-acetyl-galactosamine-iib2 component (agav, spy_0631) from streptococcus pyogenes at 1.65 a3 resolution
6 c1vsqC_
100.0
34
PDB header: transferaseChain: C: PDB Molecule: mannose-specific phosphotransferase enzyme iibPDBTitle: solution nmr structure of the productive complex between2 iiamannose and iibmannose of the mannose transporter of3 the e. coli phosphotransferase system
7 c3l0gD_
66.3
16
PDB header: transferaseChain: D: PDB Molecule: nicotinate-nucleotide pyrophosphorylase;PDBTitle: crystal structure of nicotinate-nucleotide pyrophosphorylase from2 ehrlichia chaffeensis at 2.05a resolution
8 c1qapA_
64.2
8
PDB header: glycosyltransferaseChain: A: PDB Molecule: quinolinic acid phosphoribosyltransferase;PDBTitle: quinolinic acid phosphoribosyltransferase with bound2 quinolinic acid
9 d1r8ja2
44.8
12
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: N-terminal domain of the circadian clock protein KaiA10 c3rfaA_
38.7
13
PDB header: oxidoreductaseChain: A: PDB Molecule: ribosomal rna large subunit methyltransferase n;PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
11 d1qpoa1
34.4
11
Fold: TIM beta/alpha-barrelSuperfamily: Nicotinate/Quinolinate PRTase C-terminal domain-likeFamily: NadC C-terminal domain-like12 c3pm7A_
32.2
3
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of ef_3132 protein from enterococcus faecalis at the2 resolution 2a, northeast structural genomics consortium target efr184
13 c3ilhA_
29.4
9
PDB header: transcription regulatorChain: A: PDB Molecule: two component response regulator;PDBTitle: crystal structure of two component response regulator from cytophaga2 hutchinsonii
14 c1qpoA_
28.4
11
PDB header: transferaseChain: A: PDB Molecule: quinolinate acid phosphoribosyl transferase;PDBTitle: quinolinate phosphoribosyl transferase (qaprtase) apo-enzyme from2 mycobacterium tuberculosis
15 c2k1oA_
26.8
22
PDB header: gene regulationChain: A: PDB Molecule: putative;PDBTitle: nmr structure of helicobacter pylori jhp0511 (hp0564).
16 d1k68a_
25.7
9
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related17 d1qapa1
25.3
6
Fold: TIM beta/alpha-barrelSuperfamily: Nicotinate/Quinolinate PRTase C-terminal domain-likeFamily: NadC C-terminal domain-like18 c3gl9B_
23.3
15
PDB header: signaling proteinChain: B: PDB Molecule: response regulator;PDBTitle: the structure of a histidine kinase-response regulator2 complex sheds light into two-component signaling and3 reveals a novel cis autophosphorylation mechanism
19 c3pajA_
22.5
11
PDB header: transferaseChain: A: PDB Molecule: nicotinate-nucleotide pyrophosphorylase, carboxylating;PDBTitle: 2.00 angstrom resolution crystal structure of a quinolinate2 phosphoribosyltransferase from vibrio cholerae o1 biovar eltor str.3 n16961
20 c3obhA_
20.9
4
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: x-ray crystal structure of protein sp_0782 (7-79) from streptococcus2 pneumoniae. northeast structural genomics consortium target spr104
21 c3khtA_
not modelled
19.6
10
PDB header: signaling proteinChain: A: PDB Molecule: response regulator;PDBTitle: crystal structure of response regulator from hahella chejuensis
22 d1t0kb_
not modelled
18.8
10
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: L30e/L7ae ribosomal proteins23 c2yxxA_
not modelled
18.1
16
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: crystal structure analysis of diaminopimelate decarboxylate (lysa)
24 c3eqzB_
not modelled
17.7
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: response regulator;PDBTitle: crystal structure of a response regulator from colwellia2 psychrerythraea
25 d1mb3a_
not modelled
16.3
13
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related26 c2oqrA_
not modelled
15.8
20
PDB header: transcription,signaling proteinChain: A: PDB Molecule: sensory transduction protein regx3;PDBTitle: the structure of the response regulator regx3 from mycobacterium2 tuberculosis
27 d2pl1a1
not modelled
15.2
12
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related28 c3n53B_
not modelled
14.2
9
PDB header: transcriptionChain: B: PDB Molecule: response regulator receiver modulated diguanylate cyclase;PDBTitle: crystal structure of a response regulator receiver modulated2 diguanylate cyclase from pelobacter carbinolicus
29 c2qv0A_
not modelled
14.1
11
PDB header: transcriptionChain: A: PDB Molecule: protein mrke;PDBTitle: crystal structure of the response regulatory domain of2 protein mrke from klebsiella pneumoniae
30 c3h5iA_
not modelled
13.6
13
PDB header: transcriptionChain: A: PDB Molecule: response regulator/sensory box protein/ggdefPDBTitle: crystal structure of the n-terminal domain of a response2 regulator/sensory box/ggdef 3-domain protein from3 carboxydothermus hydrogenoformans
31 d1ajza_
not modelled
13.6
19
Fold: TIM beta/alpha-barrelSuperfamily: Dihydropteroate synthetase-likeFamily: Dihydropteroate synthetase32 c2a5hC_
not modelled
13.3
15
PDB header: isomeraseChain: C: PDB Molecule: l-lysine 2,3-aminomutase;PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
33 d1k66a_
not modelled
12.8
17
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related34 c1rnlA_
not modelled
12.8
15
PDB header: signal transduction proteinChain: A: PDB Molecule: nitrate/nitrite response regulator protein narl;PDBTitle: the nitrate/nitrite response regulator protein narl from narl
35 d1kgsa1
not modelled
12.4
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: PhoB-like36 d2i7pa1
not modelled
11.8
19
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Fumble-like37 c3cnbC_
not modelled
11.7
9
PDB header: dna binding proteinChain: C: PDB Molecule: dna-binding response regulator, merr family;PDBTitle: crystal structure of signal receiver domain of dna binding response2 regulator protein (merr) from colwellia psychrerythraea 34h
38 d1bs0a_
not modelled
11.5
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like39 c2y5sA_
not modelled
11.3
16
PDB header: transferaseChain: A: PDB Molecule: dihydropteroate synthase;PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
40 d1t6t1_
not modelled
11.1
11
Fold: Toprim domainSuperfamily: Toprim domainFamily: Toprim domain41 c3btnA_
not modelled
11.0
14
PDB header: biosynthetic proteinChain: A: PDB Molecule: antizyme inhibitor 1;PDBTitle: crystal structure of antizyme inhibitor, an ornithine2 decarboxylase homologous protein
42 c3m6mF_
not modelled
10.9
18
PDB header: lyase/transferaseChain: F: PDB Molecule: sensory/regulatory protein rpfc;PDBTitle: crystal structure of rpff complexed with rec domain of rpfc
43 d2f1fa1
not modelled
10.8
13
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like44 d1a04a2
not modelled
10.7
15
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related45 c3cz5B_
not modelled
10.1
12
PDB header: transcription regulatorChain: B: PDB Molecule: two-component response regulator, luxr family;PDBTitle: crystal structure of two-component response regulator, luxr family,2 from aurantimonas sp. si85-9a1
46 c3cg4A_
not modelled
10.0
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: response regulator receiver domain protein (chey-like);PDBTitle: crystal structure of response regulator receiver domain protein (chey-2 like) from methanospirillum hungatei jf-1
47 d1ad1a_
not modelled
9.9
24
Fold: TIM beta/alpha-barrelSuperfamily: Dihydropteroate synthetase-likeFamily: Dihydropteroate synthetase48 d1dbwa_
not modelled
9.8
6
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related49 d2fgca2
not modelled
9.8
19
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like50 c3hebB_
not modelled
9.7
16
PDB header: transcription regulatorChain: B: PDB Molecule: response regulator receiver domain protein (chey);PDBTitle: crystal structure of response regulator receiver domain from2 rhodospirillum rubrum
51 d1oqya2
not modelled
9.6
18
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain52 d1u0sy_
not modelled
9.5
13
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related53 c2rjnA_
not modelled
9.5
13
PDB header: hydrolaseChain: A: PDB Molecule: response regulator receiver:metal-dependentPDBTitle: crystal structure of an uncharacterized protein q2bku2 from2 neptuniibacter caesariensis
54 d1xxaa_
not modelled
9.3
33
Fold: DCoH-likeSuperfamily: C-terminal domain of arginine repressorFamily: C-terminal domain of arginine repressor55 d2obba1
not modelled
9.3
7
Fold: HAD-likeSuperfamily: HAD-likeFamily: BT0820-like56 c3hv2B_
not modelled
9.3
6
PDB header: signaling proteinChain: B: PDB Molecule: response regulator/hd domain protein;PDBTitle: crystal structure of signal receiver domain of hd domain-2 containing protein from pseudomonas fluorescens pf-5
57 d2i7na2
not modelled
9.1
14
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Fumble-like58 d2pc6a2
not modelled
8.8
8
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like59 c3c3wB_
not modelled
8.7
15
PDB header: transcriptionChain: B: PDB Molecule: two component transcriptional regulatory protein devr;PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic response2 regulator dosr
60 c2yr1B_
not modelled
8.6
19
PDB header: lyaseChain: B: PDB Molecule: 3-dehydroquinate dehydratase;PDBTitle: crystal structure of 3-dehydroquinate dehydratase from geobacillus2 kaustophilus hta426
61 d2p5ma1
not modelled
8.5
29
Fold: DCoH-likeSuperfamily: C-terminal domain of arginine repressorFamily: C-terminal domain of arginine repressor62 c3cg0A_
not modelled
8.4
12
PDB header: lyaseChain: A: PDB Molecule: response regulator receiver modulated diguanylate cyclasePDBTitle: crystal structure of signal receiver domain of modulated diguanylate2 cyclase from desulfovibrio desulfuricans g20, an example of alternate3 folding
63 c3khdC_
not modelled
8.3
15
PDB header: transferaseChain: C: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of pff1300w.
64 c3kyiB_
not modelled
8.3
12
PDB header: transferaseChain: B: PDB Molecule: chey6 protein;PDBTitle: crystal structure of the phosphorylated p1 domain of chea3 in complex2 with chey6 from r. sphaeroides
65 c2y7eA_
not modelled
8.3
12
PDB header: lyaseChain: A: PDB Molecule: 3-keto-5-aminohexanoate cleavage enzyme;PDBTitle: crystal structure of the 3-keto-5-aminohexanoate cleavage enzyme2 (kce) from candidatus cloacamonas acidaminovorans (tetragonal form)
66 c3hzhA_
not modelled
8.1
14
PDB header: signaling proteinChain: A: PDB Molecule: chemotaxis response regulator (chey-3);PDBTitle: crystal structure of the chex-chey-bef3-mg+2 complex from2 borrelia burgdorferi
67 c2qxyB_
not modelled
8.1
18
PDB header: transcriptionChain: B: PDB Molecule: response regulator;PDBTitle: crystal structure of a response regulator from thermotoga2 maritima
68 d1b4ba_
not modelled
8.0
29
Fold: DCoH-likeSuperfamily: C-terminal domain of arginine repressorFamily: C-terminal domain of arginine repressor69 c3c3mA_
not modelled
8.0
23
PDB header: signaling proteinChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of the n-terminal domain of response regulator2 receiver protein from methanoculleus marisnigri jr1
70 c3icuA_
not modelled
8.0
16
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase rnf128;PDBTitle: protease-associated domain of the e3 ligase grail
71 c3cagF_
not modelled
7.8
29
PDB header: dna binding proteinChain: F: PDB Molecule: arginine repressor;PDBTitle: crystal structure of the oligomerization domain hexamer of the2 arginine repressor protein from mycobacterium tuberculosis in complex3 with 9 arginines.
72 d2guka1
not modelled
7.8
17
Fold: PG1857-likeSuperfamily: PG1857-likeFamily: PG1857-like73 c1a2oB_
not modelled
7.8
15
PDB header: bacterial chemotaxisChain: B: PDB Molecule: cheb methylesterase;PDBTitle: structural basis for methylesterase cheb regulation by a2 phosphorylation-activated domain
74 d1a2oa1
not modelled
7.8
14
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related75 d1o57a2
not modelled
7.7
6
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)76 c3eywA_
not modelled
7.6
19
PDB header: transport proteinChain: A: PDB Molecule: c-terminal domain of glutathione-regulated potassium-effluxPDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
77 d1i3ca_
not modelled
7.2
13
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related78 c1k3rA_
not modelled
7.2
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein mt0001;PDBTitle: crystal structure of the methyltransferase with a knot from2 methanobacterium thermoautotrophicum
79 c2zayA_
not modelled
7.2
23
PDB header: signaling proteinChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of response regulator from desulfuromonas2 acetoxidans
80 c1dd9A_
not modelled
7.0
12
PDB header: transferaseChain: A: PDB Molecule: dna primase;PDBTitle: structure of the dnag catalytic core
81 d1dd9a_
not modelled
7.0
12
Fold: DNA primase coreSuperfamily: DNA primase coreFamily: DNA primase DnaG catalytic core82 c3eulB_
not modelled
7.0
16
PDB header: transcriptionChain: B: PDB Molecule: possible nitrate/nitrite response transcriptionalPDBTitle: structure of the signal receiver domain of the putative2 response regulator narl from mycobacterium tuberculosis
83 d1dcfa_
not modelled
6.7
14
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: Receiver domain of the ethylene receptor84 c2q97T_
not modelled
6.5
20
PDB header: structural protein/cell invasionChain: T: PDB Molecule: toxofilin;PDBTitle: complex of mammalian actin with toxofilin from toxoplasma gondii
85 c3f6cB_
not modelled
6.4
9
PDB header: dna binding proteinChain: B: PDB Molecule: positive transcription regulator evga;PDBTitle: crystal structure of n-terminal domain of positive transcription2 regulator evga from escherichia coli
86 c1o57A_
not modelled
6.4
6
PDB header: dna binding proteinChain: A: PDB Molecule: pur operon repressor;PDBTitle: crystal structure of the purine operon repressor of2 bacillus subtilis
87 c3luaA_
not modelled
6.1
8
PDB header: transcription regulatorChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of a signal receiver domain of two component signal2 transduction (histidine kinase) from clostridium thermocellum
88 c3jpyA_
not modelled
6.1
15
PDB header: transport proteinChain: A: PDB Molecule: glutamate [nmda] receptor subunit epsilon-2;PDBTitle: crystal structure of the zinc-bound amino terminal domain of the nmda2 receptor subunit nr2b
89 c2b7pA_
not modelled
6.1
20
PDB header: transferaseChain: A: PDB Molecule: probable nicotinate-nucleotide pyrophosphorylase;PDBTitle: crystal structure of quinolinic acid phosphoribosyltransferase from2 helicobacter pylori
90 c2jrlA_
not modelled
6.0
18
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: solution structure of the beryllofluoride-activated ntrc4 receiver2 domain dimer
91 c3lteH_
not modelled
6.0
13
PDB header: transcriptionChain: H: PDB Molecule: response regulator;PDBTitle: crystal structure of response regulator (signal receiver domain) from2 bermanella marisrubri
92 d1mzva_
not modelled
6.0
23
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosyltransferases (PRTases)93 c2is8A_
not modelled
6.0
12
PDB header: structural proteinChain: A: PDB Molecule: molybdopterin biosynthesis enzyme, moab;PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
94 c3jteA_
not modelled
6.0
10
PDB header: protein bindingChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of response regulator receiver domain2 protein from clostridium thermocellum
95 c1b4aA_
not modelled
5.9
29
PDB header: repressorChain: A: PDB Molecule: arginine repressor;PDBTitle: structure of the arginine repressor from bacillus stearothermophilus
96 d1k3ra2
not modelled
5.8
11
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: Hypothetical protein MTH1 (MT0001), dimerisation domain97 c2hwvA_
not modelled
5.8
11
PDB header: transcriptionChain: A: PDB Molecule: dna-binding response regulator vicr;PDBTitle: crystal structure of an essential response regulator dna binding2 domain, vicrc in enterococcus faecalis, a member of the yycf3 subfamily.
98 c2k5jB_
not modelled
5.7
10
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein yiif;PDBTitle: solution structure of protein yiif from shigella flexneri2 serotype 5b (strain 8401) . northeast structural genomics3 consortium target sft1
99 d1dz3a_
not modelled
5.7
6
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related