| 1 |
|
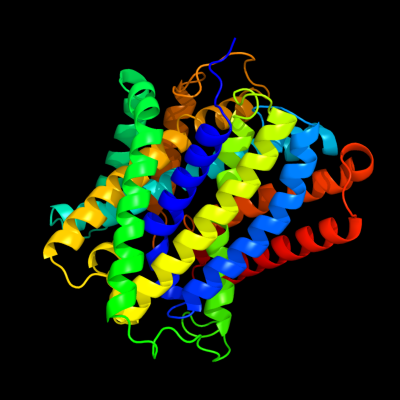
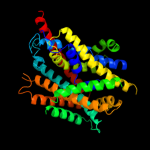
PDB 3gia chain A
Region: 5 - 411
Aligned: 399
Modelled: 407
Confidence: 100.0%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 2 |
|
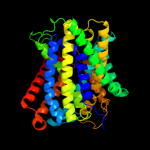
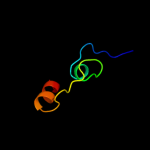
PDB 3lrc chain C
Region: 5 - 401
Aligned: 374
Modelled: 374
Confidence: 100.0%
Identity: 14%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 3 |
|
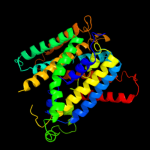
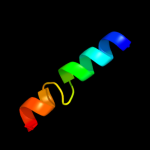
PDB 2jln chain A
Region: 2 - 413
Aligned: 406
Modelled: 410
Confidence: 99.9%
Identity: 8%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 4 |
|
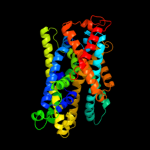
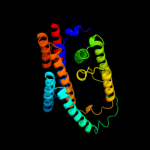
PDB 2xq2 chain A
Region: 7 - 405
Aligned: 383
Modelled: 383
Confidence: 98.8%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 5 |
|
PDB 3dh4 chain A
Region: 3 - 405
Aligned: 388
Modelled: 401
Confidence: 98.7%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 6 |
|
PDB 2a65 chain A domain 1
Region: 10 - 413
Aligned: 401
Modelled: 404
Confidence: 98.0%
Identity: 15%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 7 |
|
PDB 2w8a chain C
Region: 2 - 381
Aligned: 375
Modelled: 380
Confidence: 96.9%
Identity: 11%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 8 |
|
PDB 3hfx chain A
Region: 4 - 378
Aligned: 370
Modelled: 370
Confidence: 62.0%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 9 |
|
PDB 1llc chain A domain 1
Region: 1 - 35
Aligned: 24
Modelled: 35
Confidence: 10.7%
Identity: 25%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: LDH N-terminal domain-like
Phyre2
| 10 |
|
PDB 1s0y chain A
Region: 2 - 24
Aligned: 21
Modelled: 23
Confidence: 9.7%
Identity: 29%
Fold: Tautomerase/MIF
Superfamily: Tautomerase/MIF
Family: 4-oxalocrotonate tautomerase-like
Phyre2
| 11 |
|
PDB 1byy chain A
Region: 1 - 7
Aligned: 7
Modelled: 7
Confidence: 7.7%
Identity: 43%
PDB header:membrane protein
Chain: A: PDB Molecule:protein (sodium channel alpha-subunit);
PDBTitle: sodium channel iia inactivation gate
Phyre2
| 12 |
|
PDB 3qe7 chain A
Region: 84 - 367
Aligned: 284
Modelled: 284
Confidence: 7.5%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:uracil permease;
PDBTitle: crystal structure of uracil transporter--uraa
Phyre2
| 13 |
|
PDB 1v92 chain A
Region: 1 - 15
Aligned: 15
Modelled: 15
Confidence: 5.7%
Identity: 20%
Fold: RuvA C-terminal domain-like
Superfamily: UBA-like
Family: TAP-C domain-like
Phyre2