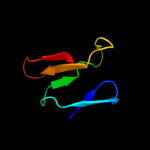
1 d1tu1a_
99.0
16
Fold: Mog1p/PsbP-likeSuperfamily: Mog1p/PsbP-likeFamily: PA0094-like2 c2xb3A_
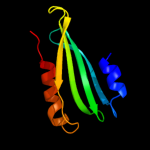
97.7
13
PDB header: photosynthesisChain: A: PDB Molecule: psbp protein;PDBTitle: the structure of cyanobacterial psbp
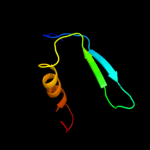
3 d1v2ba_
89.6
10
Fold: Mog1p/PsbP-likeSuperfamily: Mog1p/PsbP-likeFamily: PsbP-like4 c2vu4A_
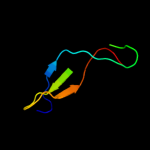
89.3
10
PDB header: photosynthesisChain: A: PDB Molecule: oxygen-evolving enhancer protein 2;PDBTitle: structure of psbp protein from spinacia oleracea at 1.98 a2 resolution
5 c2jx5A_
45.6
26
PDB header: ribosomal proteinChain: A: PDB Molecule: glub(s27a);PDBTitle: solution structure of the ubiquitin domain n-terminal to2 the s27a ribosomal subunit of giardia lamblia
6 d1qhoa3
31.1
33
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain7 d1ji1a2
30.2
17
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain8 c3orjA_
24.0
18
PDB header: sugar binding proteinChain: A: PDB Molecule: sugar-binding protein;PDBTitle: crystal structure of a sugar-binding protein (bacova_04391) from2 bacteroides ovatus at 2.16 a resolution
9 d2c7fa1
22.0
28
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: Composite domain of glycosyl hydrolase families 5, 30, 39 and 5110 d1jhsa_
21.9
23
Fold: Mog1p/PsbP-likeSuperfamily: Mog1p/PsbP-likeFamily: Ran-binding protein mog1p11 d1cgta3
20.9
33
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain12 d1qw9a1
16.9
29
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: Composite domain of glycosyl hydrolase families 5, 30, 39 and 5113 d1wzla2
16.5
33
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain14 c3lydA_
13.1
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of putative uncharacterized protein from jonesia2 denitrificans
15 c3iz5W_
12.3
30
PDB header: ribosomeChain: W: PDB Molecule: 60s ribosomal protein l22 (l22e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
16 d1xrsb2
12.3
83
Fold: Dodecin subunit-likeSuperfamily: D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domainFamily: D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domain17 d3bmva3
12.1
45
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain18 c1bagA_
10.7
28
PDB header: alpha-amylaseChain: A: PDB Molecule: alpha-1,4-glucan-4-glucanohydrolase;PDBTitle: alpha-amylase from bacillus subtilis complexed with2 maltopentaose
19 c2j98A_
10.6
13
PDB header: rna-binding proteinChain: A: PDB Molecule: replicase polyprotein 1ab;PDBTitle: human coronavirus 229e non structural protein 9 cys69ala2 mutant (nsp9)
20 c4a1dM_
9.7
27
PDB header: ribosomeChain: M: PDB Molecule: ribosomal protein l22;PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of3 molecule 4.
21 d1lmia_
not modelled
8.2
22
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Antigen MPT63/MPB63 (immunoprotective extracellular protein)Family: Antigen MPT63/MPB63 (immunoprotective extracellular protein)22 d1ni2a3
not modelled
8.2
23
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: First domain of FERM23 c3es1A_
not modelled
7.9
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: cupin 2, conserved barrel domain protein;PDBTitle: crystal structure of protein with a cupin-like fold and unknown2 function (yp_001165807.1) from novosphingobium aromaticivorans dsm3 12444 at 1.91 a resolution
24 c3mx7A_
not modelled
6.8
18
PDB header: apoptosisChain: A: PDB Molecule: fas apoptotic inhibitory molecule 1;PDBTitle: crystal structure analysis of human faim-ntd
25 d1szwa_
not modelled
6.8
44
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: tRNA pseudouridine synthase TruD26 c1bplB_
not modelled
6.6
18
PDB header: glycosyltransferaseChain: B: PDB Molecule: alpha-1,4-glucan-4-glucanohydrolase;PDBTitle: glycosyltransferase
27 c1z2zB_
not modelled
6.5
44
PDB header: lyaseChain: B: PDB Molecule: probable trna pseudouridine synthase d;PDBTitle: crystal structure of the putative trna pseudouridine2 synthase d (trud) from methanosarcina mazei, northeast3 structural genomics target mar1
28 d1s21a_
not modelled
6.4
42
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: AvrPphF ORF2, a type III effector29 c1s21A_
not modelled
6.4
42
PDB header: chaperoneChain: A: PDB Molecule: orf2;PDBTitle: crystal structure of avrpphf orf2, a type iii effector from2 p. syringae
30 c2gs8A_
not modelled
6.2
14
PDB header: lyaseChain: A: PDB Molecule: mevalonate pyrophosphate decarboxylase;PDBTitle: structure of mevalonate pyrophosphate decarboxylase from streptococcus2 pyogenes
31 c1sb7A_
not modelled
6.2
44
PDB header: lyaseChain: A: PDB Molecule: trna pseudouridine synthase d;PDBTitle: crystal structure of the e.coli pseudouridine synthase trud
32 d2bsya1
not modelled
5.6
17
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like33 d1zupa1
not modelled
5.6
28
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: TM1739-like34 c2zt9E_
not modelled
5.5
42
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex from nostoc sp. pcc2 7120
35 d1ua7a1
not modelled
5.4
26
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain36 d1x38a2
not modelled
5.3
15
Fold: Flavodoxin-likeSuperfamily: Beta-D-glucan exohydrolase, C-terminal domainFamily: Beta-D-glucan exohydrolase, C-terminal domain37 d1cxla3
not modelled
5.3
45
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain