| 1 |
|
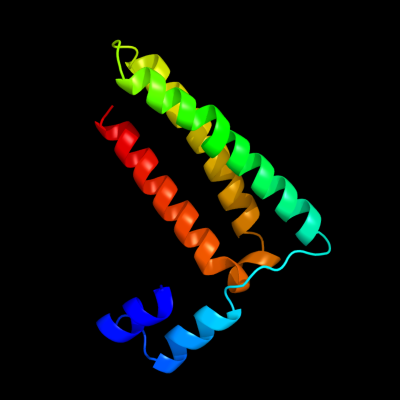
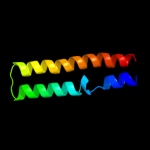
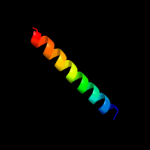
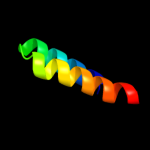
PDB 1y4c chain A
Region: 4 - 118
Aligned: 112
Modelled: 115
Confidence: 69.4%
Identity: 11%
PDB header:de novo protein
Chain: A: PDB Molecule:maltose binding protein fused with designed
PDBTitle: designed helical protein fusion mbp
Phyre2
| 2 |
|
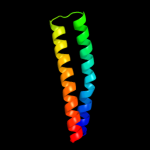
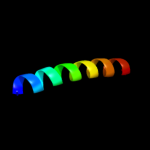
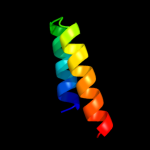
PDB 3u59 chain C
Region: 26 - 120
Aligned: 95
Modelled: 95
Confidence: 67.5%
Identity: 18%
PDB header:contractile protein
Chain: C: PDB Molecule:tropomyosin beta chain;
PDBTitle: n-terminal 98-aa fragment of smooth muscle tropomyosin beta
Phyre2
| 3 |
|
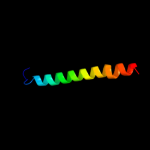
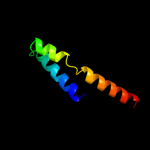
PDB 1k4t chain A domain 1
Region: 71 - 120
Aligned: 50
Modelled: 50
Confidence: 37.6%
Identity: 28%
Fold: Long alpha-hairpin
Superfamily: Eukaryotic DNA topoisomerase I, dispensable insert domain
Family: Eukaryotic DNA topoisomerase I, dispensable insert domain
Phyre2
| 4 |
|
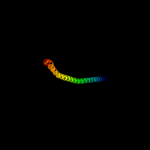
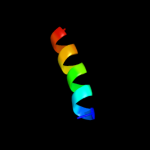
PDB 2k48 chain A
Region: 54 - 117
Aligned: 64
Modelled: 64
Confidence: 31.1%
Identity: 20%
PDB header:viral protein
Chain: A: PDB Molecule:nucleoprotein;
PDBTitle: nmr structure of the n-terminal coiled coil domain of the2 andes hantavirus nucleocapsid protein
Phyre2
| 5 |
|
PDB 2ic6 chain B
Region: 54 - 117
Aligned: 64
Modelled: 64
Confidence: 22.3%
Identity: 23%
PDB header:viral protein
Chain: B: PDB Molecule:nucleocapsid protein;
PDBTitle: the coiled-coil domain (residues 1-75) structure of the sin2 nombre virus nucleocapsid protein
Phyre2
| 6 |
|
PDB 1d7m chain A
Region: 44 - 116
Aligned: 62
Modelled: 73
Confidence: 19.3%
Identity: 26%
PDB header:contractile protein
Chain: A: PDB Molecule:cortexillin i;
PDBTitle: coiled-coil dimerization domain from cortexillin i
Phyre2
| 7 |
|
PDB 2khk chain A
Region: 67 - 106
Aligned: 39
Modelled: 40
Confidence: 17.9%
Identity: 23%
PDB header:transport protein
Chain: A: PDB Molecule:atp synthase subunit b;
PDBTitle: nmr solution structure of the b30-82 domain of subunit b of2 escherichia coli f1fo atp synthase
Phyre2
| 8 |
|
PDB 3ghg chain K
Region: 22 - 120
Aligned: 93
Modelled: 99
Confidence: 17.5%
Identity: 10%
PDB header:blood clotting
Chain: K: PDB Molecule:fibrinogen beta chain;
PDBTitle: crystal structure of human fibrinogen
Phyre2
| 9 |
|
PDB 3ipk chain A
Region: 25 - 119
Aligned: 94
Modelled: 95
Confidence: 16.4%
Identity: 17%
PDB header:cell adhesion
Chain: A: PDB Molecule:agi/ii;
PDBTitle: crystal structure of a3vp1 of agi/ii of streptococcus mutans
Phyre2
| 10 |
|
PDB 3hnw chain B
Region: 9 - 91
Aligned: 77
Modelled: 83
Confidence: 15.3%
Identity: 6%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a basic coiled-coil protein of unknown function2 from eubacterium eligens atcc 27750
Phyre2
| 11 |
|
PDB 3err chain B
Region: 71 - 122
Aligned: 51
Modelled: 52
Confidence: 14.9%
Identity: 18%
PDB header:ligase
Chain: B: PDB Molecule:fusion protein of microtubule binding domain from
PDBTitle: microtubule binding domain from mouse cytoplasmic dynein as2 a fusion with seryl-trna synthetase
Phyre2
| 12 |
|
PDB 2fxm chain B
Region: 25 - 120
Aligned: 96
Modelled: 96
Confidence: 14.0%
Identity: 14%
PDB header:contractile protein
Chain: B: PDB Molecule:myosin heavy chain, cardiac muscle beta isoform;
PDBTitle: structure of the human beta-myosin s2 fragment
Phyre2
| 13 |
|
PDB 2qdq chain A
Region: 91 - 118
Aligned: 28
Modelled: 28
Confidence: 9.9%
Identity: 25%
PDB header:structural protein
Chain: A: PDB Molecule:talin-1;
PDBTitle: crystal structure of the talin dimerisation domain
Phyre2
| 14 |
|
PDB 1zxa chain B
Region: 95 - 118
Aligned: 24
Modelled: 24
Confidence: 8.7%
Identity: 29%
PDB header:transferase
Chain: B: PDB Molecule:cgmp-dependent protein kinase 1, alpha isozyme;
PDBTitle: solution structure of the coiled-coil domain of cgmp-2 dependent protein kinase ia
Phyre2
| 15 |
|
PDB 1vp7 chain A
Region: 67 - 117
Aligned: 51
Modelled: 51
Confidence: 7.2%
Identity: 12%
Fold: Spectrin repeat-like
Superfamily: XseB-like
Family: XseB-like
Phyre2
| 16 |
|
PDB 1r48 chain A
Region: 104 - 120
Aligned: 17
Modelled: 17
Confidence: 6.8%
Identity: 29%
PDB header:transport protein
Chain: A: PDB Molecule:proline/betaine transporter;
PDBTitle: solution structure of the c-terminal cytoplasmic domain2 residues 468-497 of escherichia coli protein prop
Phyre2
| 17 |
|
PDB 2gl2 chain B
Region: 21 - 120
Aligned: 94
Modelled: 100
Confidence: 6.8%
Identity: 12%
PDB header:cell adhesion
Chain: B: PDB Molecule:adhesion a;
PDBTitle: crystal structure of the tetra muntant (t66g,r67g,f68g,2 y69g) of bacterial adhesin fada
Phyre2
| 18 |
|
PDB 3l8r chain A
Region: 83 - 117
Aligned: 35
Modelled: 35
Confidence: 6.0%
Identity: 29%
PDB header:transferase
Chain: A: PDB Molecule:putative pts system, cellobiose-specific iia
PDBTitle: the crystal structure of ptca from s. mutans
Phyre2
| 19 |
|
PDB 1wcr chain A
Region: 83 - 117
Aligned: 35
Modelled: 35
Confidence: 5.7%
Identity: 9%
PDB header:transferase
Chain: A: PDB Molecule:pts system, n, n'-diacetylchitobiose-specific
PDBTitle: trimeric structure of the enzyme iia from escherichia coli2 phosphotransferase system specific for n,n'-3 diacetylchitobiose
Phyre2
| 20 |
|
PDB 2d3e chain D
Region: 25 - 120
Aligned: 96
Modelled: 96
Confidence: 5.5%
Identity: 18%
PDB header:contractile protein
Chain: D: PDB Molecule:general control protein gcn4 and tropomyosin 1
PDBTitle: crystal structure of the c-terminal fragment of rabbit2 skeletal alpha-tropomyosin
Phyre2
| 21 |
|
| 22 |
|