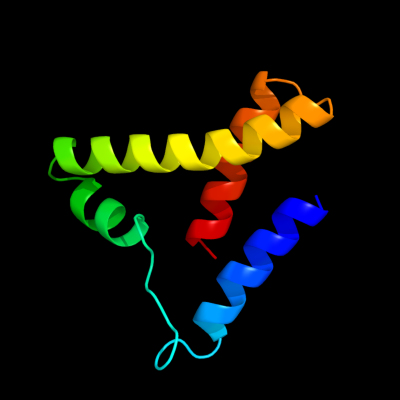
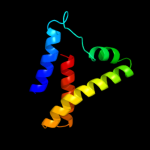
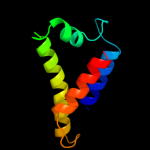
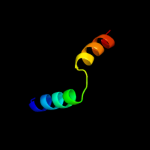
1 d1sgma2
60.0
10
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain2 d2hyja2
47.5
14
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain3 c3dfgA_
36.8
20
PDB header: recombinationChain: A: PDB Molecule: regulatory protein recx;PDBTitle: crystal structure of recx: a potent inhibitor protein of2 reca from xanthomonas campestris
4 c3c1dA_
35.6
16
PDB header: recombination, dna binding proteinChain: A: PDB Molecule: regulatory protein recx;PDBTitle: x-ray crystal structure of recx
5 d2g7sa2
26.3
15
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain6 d1uw0a_
25.4
21
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: PARP-type zinc finger7 d1ug8a_
25.2
14
Fold: IF3-likeSuperfamily: R3H domainFamily: R3H domain8 c3fgrA_
23.4
16
PDB header: hydrolaseChain: A: PDB Molecule: putative phospholipase b-like 2 28 kda form;PDBTitle: two chain form of the 66.3 kda protein at 1.8 angstroem
9 c2k0mA_
20.8
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of the uncharacterized protein from2 rhodospirillum rubrum gene locus rru_a0810. northeast3 structural genomics target rrr43
10 d1pzra_
18.8
100
Fold: HLH-likeSuperfamily: Docking domain B of the erythromycin polyketide synthase (DEBS)Family: Docking domain B of the erythromycin polyketide synthase (DEBS)11 d2i76a1
17.4
12
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: ProC C-terminal domain-like12 d1hlba_
15.9
24
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins13 d1xhoa_
14.0
17
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: Chorismate mutase14 c1xhoB_
14.0
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: chorismate mutase;PDBTitle: chorismate mutase from clostridium thermocellum cth-682
15 c2vxdA_
13.5
26
PDB header: nuclear proteinChain: A: PDB Molecule: nucleophosmin;PDBTitle: the structure of the c-terminal domain of nucleophosmin
16 d1fnja_
13.1
24
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: Chorismate mutase17 d2v4jc1
12.8
13
Fold: DsrC, the gamma subunit of dissimilatory sulfite reductaseSuperfamily: DsrC, the gamma subunit of dissimilatory sulfite reductaseFamily: DsrC, the gamma subunit of dissimilatory sulfite reductase18 c2dmjA_
12.7
38
PDB header: transferaseChain: A: PDB Molecule: poly (adp-ribose) polymerase family, member 1;PDBTitle: solution structure of the first zf-parp domain of human2 poly(adp-ribose)polymerase-1
19 d2lhba_
12.6
13
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins20 c3hugJ_
12.3
28
PDB header: transcription/membrane proteinChain: J: PDB Molecule: probable conserved membrane protein;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
21 c3s1sA_
not modelled
11.8
24
PDB header: hydrolase, transferaseChain: A: PDB Molecule: restriction endonuclease bpusi;PDBTitle: characterization and crystal structure of the type iig restriction2 endonuclease bpusi
22 d2ea9a1
not modelled
11.7
19
Fold: Profilin-likeSuperfamily: YeeU-likeFamily: YagB/YeeU/YfjZ-like23 d1it2a_
not modelled
11.4
18
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins24 c3e3vA_
not modelled
10.1
27
PDB header: recombinationChain: A: PDB Molecule: regulatory protein recx;PDBTitle: crystal structure of recx from lactobacillus salivarius
25 c3k66A_
not modelled
10.1
9
PDB header: cell adhesionChain: A: PDB Molecule: beta-amyloid-like protein;PDBTitle: x-ray crystal structure of the e2 domain of c. elegans apl-1
26 d1dbfa_
not modelled
9.9
24
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: Chorismate mutase27 d2h28a1
not modelled
9.3
24
Fold: Profilin-likeSuperfamily: YeeU-likeFamily: YagB/YeeU/YfjZ-like28 d2ox6a1
not modelled
9.1
33
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: YdiL-like29 c2xvcA_
not modelled
9.0
33
PDB header: cell cycleChain: A: PDB Molecule: escrt-iii;PDBTitle: molecular and structural basis of escrt-iii recruitment to2 membranes during archaeal cell division
30 d2inwa1
not modelled
8.9
24
Fold: Profilin-likeSuperfamily: YeeU-likeFamily: YagB/YeeU/YfjZ-like31 d1uc3a_
not modelled
8.5
13
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins32 d3sdha_
not modelled
8.3
25
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins33 c1o06A_
not modelled
7.1
23
PDB header: transport proteinChain: A: PDB Molecule: vacuolar protein sorting-associated protein vps27;PDBTitle: crystal structure of the vps27p ubiquitin interacting motif (uim)
34 c1q15A_
not modelled
7.1
56
PDB header: biosynthetic proteinChain: A: PDB Molecule: cara;PDBTitle: carbapenam synthetase
35 c1g2cN_
not modelled
7.1
37
PDB header: viral proteinChain: N: PDB Molecule: fusion protein (f);PDBTitle: human respiratory syncytial virus fusion protein core
36 c3mjhD_
not modelled
6.5
63
PDB header: protein transportChain: D: PDB Molecule: early endosome antigen 1;PDBTitle: crystal structure of human rab5a in complex with the c2h2 zinc finger2 of eea1
37 c3b5nE_
not modelled
6.3
15
PDB header: membrane proteinChain: E: PDB Molecule: synaptobrevin homolog 1;PDBTitle: structure of the yeast plasma membrane snare complex
38 c3iz5t_
not modelled
6.2
33
PDB header: ribosomeChain: T: PDB Molecule: 60s ribosomal protein l19 (l19e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
39 d1hlma_
not modelled
6.1
33
Fold: Globin-likeSuperfamily: Globin-likeFamily: Globins40 d2fq4a2
not modelled
6.0
36
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain41 c2vsgB_
not modelled
6.0
20
PDB header: membrane proteinChain: B: PDB Molecule: variant surface glycoprotein iltat 1.24;PDBTitle: a structural motif in the variant surface glycoproteins of2 trypanosoma brucei
42 c1o1nA_
not modelled
5.9
19
PDB header: oxygen storage/transportChain: A: PDB Molecule: hemoglobin alpha chain;PDBTitle: deoxy hemoglobin (a-glyglygly-c:v1m,l29w; b,d:v1m)
43 c1yx3A_
not modelled
5.9
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein dsrc;PDBTitle: nmr structure of allochromatium vinosum dsrc: northeast2 structural genomics consortium target op4
44 d1gdta1
not modelled
5.9
36
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain45 c2lbfA_
not modelled
5.8
42
PDB header: ribosomal proteinChain: A: PDB Molecule: 60s acidic ribosomal protein p1;PDBTitle: solution structure of the dimerization domain of human ribosomal2 protein p1/p2 heterodimer
46 c3izct_
not modelled
5.5
25
PDB header: ribosomeChain: T: PDB Molecule: 60s ribosomal protein rpl19 (l19e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
47 c2v3sB_
not modelled
5.3
21
PDB header: transferaseChain: B: PDB Molecule: serine/threonine-protein kinase osr1;PDBTitle: structural insights into the recognition of substrates and2 activators by the osr1 kinase
48 c3hq2A_
not modelled
5.2
30
PDB header: hydrolaseChain: A: PDB Molecule: bacillus subtilis m32 carboxypeptidase;PDBTitle: bsucp crystal structure