1 c2z4hB_
100.0
100
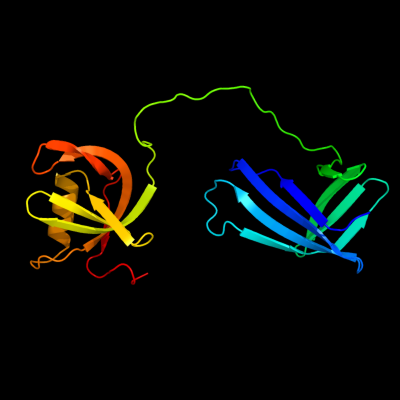
PDB header: signaling protein activatorChain: B: PDB Molecule: copper homeostasis protein cutf;PDBTitle: crystal structure of the cpx pathway activator nlpe from2 escherichia coli
2 c3lhnB_
100.0
36
PDB header: lipid binding proteinChain: B: PDB Molecule: lipoprotein;PDBTitle: crystal structure of putative lipoprotein (np_718719.1) from2 shewanella oneidensis at 1.42 a resolution
3 d1jiwi_
74.1
16
Fold: Streptavidin-likeSuperfamily: beta-Barrel protease inhibitorsFamily: Metalloprotease inhibitor4 c3ge2A_
65.8
16
PDB header: lipoproteinChain: A: PDB Molecule: lipoprotein, putative;PDBTitle: crystal structure of putative lipoprotein sp_0198 from streptococcus2 pneumoniae
5 c3p02A_
34.4
8
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein of unknown function (bacova_00267) from2 bacteroides ovatus at 1.55 a resolution
6 d2q4ma1
27.2
29
Fold: Tubby C-terminal domain-likeSuperfamily: Tubby C-terminal domain-likeFamily: At5g01750-like7 c1zxuA_
27.2
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: at5g01750 protein;PDBTitle: x-ray structure of protein from arabidopsis thaliana2 at5g01750
8 c3o0rC_
22.9
11
PDB header: immune system/oxidoreductaseChain: C: PDB Molecule: nitric oxide reductase subunit c;PDBTitle: crystal structure of nitric oxide reductase from pseudomonas2 aeruginosa in complex with antibody fragment
9 c1brvA_
18.2
47
PDB header: glycoproteinChain: A: PDB Molecule: protein g;PDBTitle: solution nmr structure of the immunodominant region of2 protein g of bovine respiratory syncytial virus, 483 structures
10 d1jhna4
18.0
21
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Calnexin/calreticulin11 d1hxra_
17.6
86
Fold: Mss4-likeSuperfamily: Mss4-likeFamily: RabGEF Mss412 c3p43A_
17.4
19
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: structure and activities of archaeal members of the ligd 3'2 phosphoesterase dna repair enzyme superfamily
13 d2fu5a1
17.4
86
Fold: Mss4-likeSuperfamily: Mss4-likeFamily: RabGEF Mss414 d1rc9a2
15.3
57
Fold: Crisp domain-likeSuperfamily: Crisp domain-likeFamily: Crisp domain15 c1nuiA_
15.3
24
PDB header: replicationChain: A: PDB Molecule: dna primase/helicase;PDBTitle: crystal structure of the primase fragment of bacteriophage t7 primase-2 helicase protein
16 d2fm8a1
15.2
17
Fold: Secretion chaperone-likeSuperfamily: Type III secretory system chaperone-likeFamily: Type III secretory system chaperone17 d2hh8a1
15.1
6
Fold: YdfO-likeSuperfamily: YdfO-likeFamily: YdfO-like18 c2l6nA_
14.1
43
PDB header: structure genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein yp_001092504.1;PDBTitle: nmr solution structure of the protein yp_001092504.1
19 c3hyxC_
13.9
56
PDB header: oxidoreductaseChain: C: PDB Molecule: sulfide-quinone reductase;PDBTitle: 3-d x-ray structure of the sulfide:quinone oxidoreductase from aquifex2 aeolicus in complex with aurachin c
20 c3rg0A_
13.8
21
PDB header: chaperoneChain: A: PDB Molecule: calreticulin;PDBTitle: structural and functional relationships between the lectin and arm2 domains of calreticulin
21 c2bx9J_
not modelled
12.9
38
PDB header: transcription regulationChain: J: PDB Molecule: tryptophan rna-binding attenuator protein-inhibitoryPDBTitle: crystal structure of b.subtilis anti-trap protein, an2 antagonist of trap-rna interactions
22 d1j8ra_
not modelled
12.6
16
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: PapG adhesin receptor-binding domain23 c2l6pA_
not modelled
11.5
43
PDB header: structure genomics, unknown functionChain: A: PDB Molecule: phac1, phac2 and phad genes;PDBTitle: nmr solution structure of the protein np_253742.1
24 c2ktsA_
not modelled
10.8
25
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein hslj;PDBTitle: nmr structure of the protein np_415897.1
25 c1jhnA_
not modelled
10.4
21
PDB header: chaperoneChain: A: PDB Molecule: calnexin;PDBTitle: crystal structure of the lumenal domain of calnexin
26 d1oyvi_
not modelled
10.0
29
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors27 d1r7aa1
not modelled
9.9
30
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain28 c2ovsB_
not modelled
9.8
8
PDB header: gene regulation, ligand binding proteinChain: B: PDB Molecule: l0044;PDBTitle: crystal strcuture of a type three secretion system protein
29 d2eppa1
not modelled
9.2
45
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H230 d2dsya1
not modelled
8.8
24
Fold: TTHA1013/TTHA0281-likeSuperfamily: TTHA1013/TTHA0281-likeFamily: TTHA0281-like31 c2dcrA_
not modelled
8.2
18
PDB header: transferaseChain: A: PDB Molecule: proto-oncogene tyrosine-protein kinase fes/fps;PDBTitle: fully automated solution structure determination of the fes2 sh2 domain
32 c3kg9A_
not modelled
8.1
11
PDB header: lyaseChain: A: PDB Molecule: curk;PDBTitle: dehydratase domain from curk module of curacin polyketide synthase
33 d1vlya1
not modelled
8.1
16
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: Aminomethyltransferase beta-barrel domainFamily: Aminomethyltransferase beta-barrel domain34 d2aqaa1
not modelled
8.1
40
Fold: Rubredoxin-likeSuperfamily: Nop10-like SnoRNPFamily: Nucleolar RNA-binding protein Nop10-like35 d2bpa2_
not modelled
8.0
29
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: ssDNA virusesFamily: Microviridae-like VP36 c3luuA_
not modelled
7.9
60
PDB header: biosynthetic proteinChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein with unknown function which belongs to2 pfam duf971 family (afe_2189) from acidithiobacillus ferrooxidans3 atcc 23270 at 1.93 a resolution
37 d1w0na_
not modelled
7.9
25
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 36 carbohydrate binding module, CBM3638 d1o6da_
not modelled
7.9
24
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: YbeA-like39 d1wgea1
not modelled
7.9
38
Fold: Rubredoxin-likeSuperfamily: CSL zinc fingerFamily: CSL zinc finger40 d1gff2_
not modelled
7.8
36
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: ssDNA virusesFamily: Microviridae-like VP41 c1n1uA_
not modelled
7.6
25
PDB header: antibioticChain: A: PDB Molecule: kalata b1;PDBTitle: nmr structure of [ala1,15]kalata b1
42 d1n1ua_
not modelled
7.6
25
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: CyclotidesFamily: Kalata B143 c1r8oA_
not modelled
7.6
38
PDB header: hydrolase inhibitorChain: A: PDB Molecule: kunitz trypsin inhibitor;PDBTitle: crystal structure of an unusual kunitz-type trypsin inhibitor from2 copaifera langsdorffii seeds
44 c3cjtP_
not modelled
7.5
6
PDB header: transferase/ribosomal proteinChain: P: PDB Molecule: 50s ribosomal protein l11;PDBTitle: ribosomal protein l11 methyltransferase (prma) in complex with2 dimethylated ribosomal protein l11
45 c3ld0Q_
not modelled
7.4
38
PDB header: gene regulationChain: Q: PDB Molecule: inhibitor of trap, regulated by t-box (trp) sequence rtpa;PDBTitle: crystal structure of b.licheniformis anti-trap protein, an antagonist2 of trap-rna interactions
46 d2gvia2
not modelled
7.4
67
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: FwdE C-terminal domain-like47 c3n9dA_
not modelled
7.3
17
PDB header: ligaseChain: A: PDB Molecule: probable atp-dependent dna ligase;PDBTitle: monoclinic structure of p. aeruginosa ligd phosphoesterase domain
48 d1mila_
not modelled
7.2
25
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain49 d1ns5a_
not modelled
7.2
24
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: YbeA-like50 c2knnA_
not modelled
7.2
50
PDB header: plant proteinChain: A: PDB Molecule: cycloviolacin-o2;PDBTitle: solution structure of the cyclotide cycloviolacin o2 with2 glu6 methylated (cyo2me)
51 c2qn4B_
not modelled
7.2
29
PDB header: hydrolase inhibitorChain: B: PDB Molecule: alpha-amylase/subtilisin inhibitor;PDBTitle: structure and function study of rice bifunctional alpha-2 amylase/subtilisin inhibitor from oryza sativa
52 d1to0a_
not modelled
7.1
24
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: YbeA-like53 d1vh0a_
not modelled
7.1
18
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: YbeA-like54 c2f2iA_
not modelled
6.8
25
PDB header: antimicrobial proteinChain: A: PDB Molecule: kalata-b1;PDBTitle: solution structure of [p20d,v21k]-kalata b1
55 d1avwb_
not modelled
6.7
57
Fold: beta-TrefoilSuperfamily: STI-likeFamily: Kunitz (STI) inhibitors56 d1nb1a_
not modelled
6.6
25
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: CyclotidesFamily: Kalata B157 d1ha8a_
not modelled
6.6
63
Fold: Pheromone ER-23Superfamily: Pheromone ER-23Family: Pheromone ER-2358 d1lrva_
not modelled
6.6
38
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: Leucine-rich repeat variant59 c3o6uB_
not modelled
6.6
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein cpe2226;PDBTitle: crystal structure of cpe2226 protein from clostridium perfringens.2 northeast structural genomics consortium target cpr195
60 d1fnoa3
not modelled
6.5
9
Fold: Ferredoxin-likeSuperfamily: Bacterial exopeptidase dimerisation domainFamily: Bacterial exopeptidase dimerisation domain61 d1pt4a_
not modelled
6.5
25
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: CyclotidesFamily: Kalata B162 c2l6lA_
not modelled
6.5
18
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily c member 24;PDBTitle: solution structure of human j-protein co-chaperone, dph4
63 c2kzxA_
not modelled
6.5
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of a3dht5 from clostridium thermocellum,2 northeast structural genomics consortium target cmr116
64 d1cf1a1
not modelled
6.4
29
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Arrestin/Vps26-like65 c2f2jA_
not modelled
6.4
25
PDB header: antimicrobial proteinChain: A: PDB Molecule: kalata-b1;PDBTitle: solution structure of [w19k, p20n, v21k]-kalata b1
66 c2zkrk_
not modelled
6.3
9
PDB header: ribosomal protein/rnaChain: K: PDB Molecule: rna expansion segment es19;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
67 c2khbA_
not modelled
6.3
25
PDB header: antimicrobial proteinChain: A: PDB Molecule: kalata-b1;PDBTitle: solution structure of linear kalata b1 (loop 6)
68 c1iebD_
not modelled
6.0
20
PDB header: histocompatibility antigenChain: D: PDB Molecule: mhc class ii i-ek;PDBTitle: histocompatibility antigen
69 d1m3va1
not modelled
6.0
57
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain70 d1r8na_
not modelled
5.8
36
Fold: beta-TrefoilSuperfamily: STI-likeFamily: Kunitz (STI) inhibitors71 d1mmsa2
not modelled
5.7
6
Fold: Ribosomal L11/L12e N-terminal domainSuperfamily: Ribosomal L11/L12e N-terminal domainFamily: Ribosomal L11/L12e N-terminal domain72 c2xvoB_
not modelled
5.7
32
PDB header: structural genomicsChain: B: PDB Molecule: sso1725;PDBTitle: sso1725, a protein involved in the crispr/cas pathway
73 d1vqok1
not modelled
5.7
14
Fold: Ribosomal protein L14Superfamily: Ribosomal protein L14Family: Ribosomal protein L1474 c3swrA_
not modelled
5.6
14
PDB header: transferaseChain: A: PDB Molecule: dna (cytosine-5)-methyltransferase 1;PDBTitle: structure of human dnmt1 (601-1600) in complex with sinefungin
75 d3cjsb1
not modelled
5.6
6
Fold: Ribosomal L11/L12e N-terminal domainSuperfamily: Ribosomal L11/L12e N-terminal domainFamily: Ribosomal L11/L12e N-terminal domain76 d1v1qa_
not modelled
5.5
14
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB77 d2hz5a1
not modelled
5.4
26
Fold: Profilin-likeSuperfamily: Roadblock/LC7 domainFamily: Roadblock/LC7 domain78 c2jmbA_
not modelled
5.4
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein atu4866;PDBTitle: solution structure of the protein atu4866 from agrobacterium2 tumefaciens
79 d1whia_
not modelled
5.4
23
Fold: Ribosomal protein L14Superfamily: Ribosomal protein L14Family: Ribosomal protein L1480 d1sg1x2
not modelled
5.3
31
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: TNF receptor-like81 c2p57A_
not modelled
5.3
55
PDB header: metal binding proteinChain: A: PDB Molecule: gtpase-activating protein znf289;PDBTitle: gap domain of znf289, an id1-regulated zinc finger protein
82 c2p3yA_
not modelled
5.3
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein vpa0735;PDBTitle: crystal structure of vpa0735 from vibrio parahaemolyticus. northeast2 structural genomics target vpr109
83 d2p3ya1
not modelled
5.3
25
Fold: VPA0735-likeSuperfamily: VPA0735-likeFamily: VPA0735-like84 d2f2ha1
not modelled
5.3
27
Fold: Putative glucosidase YicI, C-terminal domainSuperfamily: Putative glucosidase YicI, C-terminal domainFamily: Putative glucosidase YicI, C-terminal domain85 d1tiea_
not modelled
5.3
29
Fold: beta-TrefoilSuperfamily: STI-likeFamily: Kunitz (STI) inhibitors86 d1eyla_
not modelled
5.3
43
Fold: beta-TrefoilSuperfamily: STI-likeFamily: Kunitz (STI) inhibitors87 c3g12A_
not modelled
5.2
38
PDB header: lyaseChain: A: PDB Molecule: putative lactoylglutathione lyase;PDBTitle: crystal structure of a putative lactoylglutathione lyase2 from bdellovibrio bacteriovorus
88 d3bx1c1
not modelled
5.1
29
Fold: beta-TrefoilSuperfamily: STI-likeFamily: Kunitz (STI) inhibitors89 c1y7jA_
not modelled
5.1
80
PDB header: signaling proteinChain: A: PDB Molecule: agouti signaling protein;PDBTitle: nmr structure family of human agouti signalling protein (80-2 132: q115y, s124y)
90 c2owaB_
not modelled
5.1
27
PDB header: protein transportChain: B: PDB Molecule: arfgap-like finger domain containing protein;PDBTitle: crystal structure of putative gtpase activating protein for2 adp ribosylation factor from cryptosporidium parvum3 (cgd5_1040)
91 c2dreA_
not modelled
5.1
36
PDB header: plant proteinChain: A: PDB Molecule: water-soluble chlorophyll protein;PDBTitle: crystal structure of water-soluble chlorophyll protein from2 lepidium virginicum at 2.00 angstrom resolution