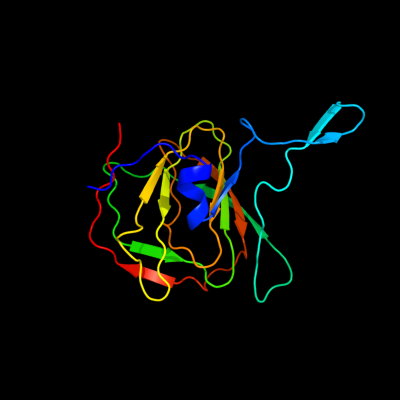
| 1 | d2bdra1
|
|
 |
100.0 |
51 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Ureidoglycolate hydrolase AllA |
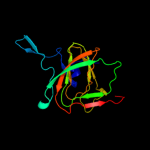
| 2 | d1xsqa_
|
|
 |
100.0 |
95 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Ureidoglycolate hydrolase AllA |
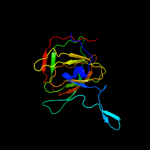
| 3 | c3d0jA_
|
|
 |
98.2 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein ca_c3497;
PDBTitle: crystal structure of conserved protein of unknown function ca_c34972 from clostridium acetobutylicum atcc 824
|
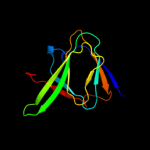
| 4 | c3mpbA_
|
|
 |
90.9 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar isomerase;
PDBTitle: z5688 from e. coli o157:h7 bound to fructose
|
| 5 | c2oa2A_
|
|
 |
89.7 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:bh2720 protein;
PDBTitle: crystal structure of bh2720 (10175341) from bacillus halodurans at2 1.41 a resolution
|
| 6 | c3kgzA_
|
|
 |
88.6 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:cupin 2 conserved barrel domain protein;
PDBTitle: crystal structure of a cupin 2 conserved barrel domain protein from2 rhodopseudomonas palustris
|
| 7 | c3jzvA_
|
|
 |
88.6 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein rru_a2000;
PDBTitle: crystal structure of rru_a2000 from rhodospirillum rubrum: a cupin-22 domain.
|
| 8 | d1nxma_
|
|
 |
88.5 |
12 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:dTDP-sugar isomerase |
| 9 | d2phla2
|
|
 |
86.7 |
13 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
| 10 | d1zvfa1
|
|
 |
85.9 |
17 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:3-hydroxyanthranilic acid dioxygenase-like |
| 11 | d2pa7a1
|
|
 |
85.2 |
13 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:dTDP-sugar isomerase |
| 12 | c2i45C_
|
|
 |
85.2 |
13 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of protein nmb1881 from neisseria meningitidis
|
| 13 | d1v70a_
|
|
 |
84.6 |
15 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:TM1287-like |
| 14 | c3ejkA_
|
|
 |
81.1 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:dtdp sugar isomerase;
PDBTitle: crystal structure of dtdp sugar isomerase (yp_390184.1) from2 desulfovibrio desulfuricans g20 at 1.95 a resolution
|
| 15 | d1o4ta_
|
|
 |
78.8 |
18 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:TM1287-like |
| 16 | c2oziA_
|
|
 |
76.6 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein rpa4178;
PDBTitle: structural genomics, the crystal structure of a putative2 protein rpa4178 from rhodopseudomonas palustris cga009
|
| 17 | c2y0oA_
|
|
 |
73.1 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:probable d-lyxose ketol-isomerase;
PDBTitle: the structure of a d-lyxose isomerase from the sigmab2 regulon of bacillus subtilis
|
| 18 | c3cewA_
|
|
 |
65.7 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized cupin protein;
PDBTitle: crystal structure of a cupin protein (bf4112) from bacteroides2 fragilis. northeast structural genomics consortium target bfr205
|
| 19 | c2gu9B_
|
|
 |
64.6 |
14 |
PDB header:immune system
Chain: B: PDB Molecule:tetracenomycin polyketide synthesis protein;
PDBTitle: crystal structure of xc5357 from xanthomonas campestris: a2 putative tetracenomycin polyketide synthesis protein3 adopting a novel cupin subfamily structure
|
| 20 | c3d82A_
|
|
 |
63.8 |
22 |
PDB header:metal binding protein
Chain: A: PDB Molecule:cupin 2, conserved barrel domain protein;
PDBTitle: crystal structure of a cupin-2 domain containing protein (sfri_3543)2 from shewanella frigidimarina ncimb 400 at 2.05 a resolution
|
| 21 | c2fqpD_ |
|
not modelled |
61.7 |
11 |
PDB header:metal binding protein
Chain: D: PDB Molecule:hypothetical protein bp2299;
PDBTitle: crystal structure of a cupin domain (bp2299) from bordetella pertussis2 tohama i at 1.80 a resolution
|
| 22 | c2vqaC_ |
|
not modelled |
61.0 |
12 |
PDB header:metal-binding protein
Chain: C: PDB Molecule:sll1358 protein;
PDBTitle: protein-folding location can regulate mn versus cu- or zn-2 binding. crystal structure of mnca.
|
| 23 | d2et1a1 |
|
not modelled |
60.4 |
22 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
| 24 | c3h8uA_ |
|
not modelled |
59.7 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized conserved protein with double-stranded
PDBTitle: crystal structure of uncharacterized conserved protein with double-2 stranded beta-helix domain (yp_001338853.1) from klebsiella3 pneumoniae subsp. pneumoniae mgh 78578 at 1.80 a resolution
|
| 25 | d1juha_ |
|
not modelled |
59.6 |
18 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Quercetin 2,3-dioxygenase-like |
| 26 | d1ep0a_ |
|
not modelled |
53.5 |
13 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:dTDP-sugar isomerase |
| 27 | d1wlta1 |
|
not modelled |
49.6 |
13 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:dTDP-sugar isomerase |
| 28 | c2o8qA_ |
|
not modelled |
45.7 |
18 |
PDB header:metal binding protein
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a protein with a cupin-like fold and unknown2 function (bxe_c0505) from burkholderia xenovorans lb400 at 1.55 a3 resolution
|
| 29 | c1cauB_ |
|
not modelled |
43.2 |
12 |
PDB header:seed storage protein
Chain: B: PDB Molecule:canavalin;
PDBTitle: determination of three crystal structures of canavalin by molecular2 replacement
|
| 30 | d2ixha1 |
|
not modelled |
39.6 |
16 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:dTDP-sugar isomerase |
| 31 | d2f4pa1 |
|
not modelled |
36.3 |
19 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:TM1287-like |
| 32 | d1vj2a_ |
|
not modelled |
35.9 |
13 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:TM1459-like |
| 33 | d1j3pa_ |
|
not modelled |
35.5 |
12 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Glucose-6-phosphate isomerase, GPI |
| 34 | c3ibmB_ |
|
not modelled |
35.3 |
21 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:cupin 2, conserved barrel domain protein;
PDBTitle: crystal structure of cupin 2 domain-containing protein hhal_0468 from2 halorhodospira halophila
|
| 35 | d2cupa1 |
|
not modelled |
34.8 |
50 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 36 | d1x82a_ |
|
not modelled |
34.0 |
12 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Glucose-6-phosphate isomerase, GPI |
| 37 | d1x63a1 |
|
not modelled |
33.6 |
50 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 38 | c2c0zA_ |
|
not modelled |
30.4 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:novw;
PDBTitle: the 1.6 a resolution crystal structure of novw: a 4-keto-6-2 deoxy sugar epimerase from the novobiocin biosynthetic3 gene cluster of streptomyces spheroides
|
| 39 | d1rc6a_ |
|
not modelled |
29.0 |
10 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:YlbA-like |
| 40 | c2cauA_ |
|
not modelled |
28.0 |
11 |
PDB header:plant protein
Chain: A: PDB Molecule:protein (canavalin);
PDBTitle: canavalin from jack bean
|
| 41 | d2fcta1 |
|
not modelled |
26.3 |
15 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:PhyH-like |
| 42 | d1y3ta1 |
|
not modelled |
25.4 |
10 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Quercetin 2,3-dioxygenase-like |
| 43 | c3l2hD_ |
|
not modelled |
25.2 |
17 |
PDB header:isomerase
Chain: D: PDB Molecule:putative sugar phosphate isomerase;
PDBTitle: crystal structure of putative sugar phosphate isomerase (afe_0303)2 from acidithiobacillus ferrooxidans atcc 23270 at 1.85 a resolution
|
| 44 | d2ixca1 |
|
not modelled |
23.5 |
15 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:dTDP-sugar isomerase |
| 45 | d1ukwa2 |
|
not modelled |
21.5 |
21 |
Fold:Acyl-CoA dehydrogenase NM domain-like
Superfamily:Acyl-CoA dehydrogenase NM domain-like
Family:Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains |
| 46 | c3chxE_ |
|
not modelled |
20.7 |
16 |
PDB header:membrane protein
Chain: E: PDB Molecule:pmob;
PDBTitle: crystal structure of methylosinus trichosporium ob3b2 particulate methane monooxygenase (pmmo)
|
| 47 | d1j58a_ |
|
not modelled |
20.5 |
15 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
| 48 | d1uija1 |
|
not modelled |
20.1 |
13 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
| 49 | c3rgbA_ |
|
not modelled |
18.9 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methane monooxygenase subunit b2;
PDBTitle: crystal structure of particulate methane monooxygenase from2 methylococcus capsulatus (bath)
|
| 50 | c3rfrI_ |
|
not modelled |
17.9 |
16 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:pmob;
PDBTitle: crystal structure of particulate methane monooxygenase (pmmo) from2 methylocystis sp. strain m
|
| 51 | d1oi6a_ |
|
not modelled |
17.2 |
10 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:dTDP-sugar isomerase |
| 52 | c3ht2A_ |
|
not modelled |
16.4 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:remf protein;
PDBTitle: zink containing polyketide cyclase remf from streptomyces2 resistomycificus
|
| 53 | d1fxza2 |
|
not modelled |
16.3 |
8 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
| 54 | d1lr5a_ |
|
not modelled |
15.9 |
12 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
| 55 | d2c0za1 |
|
not modelled |
15.6 |
14 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:dTDP-sugar isomerase |
| 56 | c3rykB_ |
|
not modelled |
14.5 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:dtdp-4-dehydrorhamnose 3,5-epimerase;
PDBTitle: 1.63 angstrom resolution crystal structure of dtdp-4-dehydrorhamnose2 3,5-epimerase (rfbc) from bacillus anthracis str. ames with tdp and3 ppi bound
|
| 57 | d1vyqa1 |
|
not modelled |
14.2 |
38 |
Fold:beta-clip
Superfamily:dUTPase-like
Family:dUTPase-like |
| 58 | c1yewI_ |
|
not modelled |
13.7 |
29 |
PDB header:oxidoreductase, membrane protein
Chain: I: PDB Molecule:particulate methane monooxygenase, b subunit;
PDBTitle: crystal structure of particulate methane monooxygenase
|
| 59 | c3i7dB_ |
|
not modelled |
12.8 |
8 |
PDB header:isomerase
Chain: B: PDB Molecule:sugar phosphate isomerase;
PDBTitle: crystal structure of sugar phosphate isomerase from a cupin2 superfamily spo2919 from silicibacter pomeroyi (yp_168127.1) from3 silicibacter pomeroyi dss-3 at 2.30 a resolution
|
| 60 | c1sefA_ |
|
not modelled |
12.6 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein;
PDBTitle: crystal structure of cupin domain protein ef2996 from enterococcus2 faecalis
|
| 61 | d1sefa_ |
|
not modelled |
12.6 |
9 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:YlbA-like |
| 62 | c3nnlB_ |
|
not modelled |
12.6 |
13 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:cura;
PDBTitle: halogenase domain from cura module (crystal form iii)
|
| 63 | c2xdvA_ |
|
not modelled |
11.8 |
16 |
PDB header:nuclear protein
Chain: A: PDB Molecule:myc-induced nuclear antigen;
PDBTitle: crystal structure of the catalytic domain of flj14393
|
| 64 | d2b8ma1 |
|
not modelled |
11.8 |
16 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:MJ0764-like |
| 65 | d1to3a_ |
|
not modelled |
11.5 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 66 | c2xxzA_ |
|
not modelled |
11.4 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lysine-specific demethylase 6b;
PDBTitle: crystal structure of the human jmjd3 jumonji domain
|
| 67 | d2jzaa1 |
|
not modelled |
11.3 |
14 |
Fold:ISP domain
Superfamily:ISP domain
Family:NirD-like |
| 68 | d1dgwa_ |
|
not modelled |
10.9 |
14 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
| 69 | d1o7na1 |
|
not modelled |
10.9 |
3 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 70 | d1oiha_ |
|
not modelled |
10.9 |
29 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:TauD/TfdA-like |
| 71 | d2jo6a1 |
|
not modelled |
10.7 |
17 |
Fold:ISP domain
Superfamily:ISP domain
Family:NirD-like |
| 72 | c1upiA_ |
|
not modelled |
10.6 |
13 |
PDB header:epimerase
Chain: A: PDB Molecule:dtdp-4-dehydrorhamnose 3,5-epimerase;
PDBTitle: mycobacterium tuberculosis rmlc epimerase (rv3465)
|
| 73 | d2cupa2 |
|
not modelled |
10.5 |
25 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 74 | d1vrba1 |
|
not modelled |
10.2 |
16 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Asparaginyl hydroxylase-like |
| 75 | d1dzra_ |
|
not modelled |
9.6 |
16 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:dTDP-sugar isomerase |
| 76 | c3fjsC_ |
|
not modelled |
9.6 |
13 |
PDB header:biosynthetic protein
Chain: C: PDB Molecule:uncharacterized protein with rmlc-like cupin fold;
PDBTitle: crystal structure of a putative biosynthetic protein with rmlc-like2 cupin fold (reut_b4087) from ralstonia eutropha jmp134 at 1.90 a3 resolution
|
| 77 | d1x4da1 |
|
not modelled |
9.2 |
9 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
| 78 | c3h7yA_ |
|
not modelled |
8.7 |
11 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:bacilysin biosynthesis protein bacb;
PDBTitle: crystal structure of bacb, an enzyme involved in bacilysin synthesis,2 in tetragonal form
|
| 79 | c2vpvA_ |
|
not modelled |
8.5 |
12 |
PDB header:cell cycle
Chain: A: PDB Molecule:protein mif2;
PDBTitle: dimerization domain of mif2p
|
| 80 | c1za4A_ |
|
not modelled |
8.0 |
19 |
PDB header:cell adhesion
Chain: A: PDB Molecule:thrombospondin 1;
PDBTitle: crystal structure of the thrombospondin-1 n-terminal domain2 in complex with arixtra
|
| 81 | c3ic3C_ |
|
not modelled |
8.0 |
35 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative pyruvate dehydrogenase;
PDBTitle: structure of a putative pyruvate dehydrogenase from the photosynthetic2 bacterium rhodopseudomonas palustrus cga009
|
| 82 | c2d40C_ |
|
not modelled |
7.8 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative gentisate 1,2-dioxygenase;
PDBTitle: crystal structure of z3393 from escherichia coli o157:h7
|
| 83 | c2d9oA_ |
|
not modelled |
7.7 |
25 |
PDB header:rna binding protein
Chain: A: PDB Molecule:dnaj (hsp40) homolog, subfamily c, member 17;
PDBTitle: solution structure of rna binding domain in hypothetical2 protein flj10634
|
| 84 | c3es1A_ |
|
not modelled |
7.7 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:cupin 2, conserved barrel domain protein;
PDBTitle: crystal structure of protein with a cupin-like fold and unknown2 function (yp_001165807.1) from novosphingobium aromaticivorans dsm3 12444 at 1.91 a resolution
|
| 85 | d1uika2 |
|
not modelled |
7.7 |
9 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
| 86 | d2erfa1 |
|
not modelled |
7.1 |
19 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Laminin G-like module |
| 87 | d1yfua1 |
|
not modelled |
7.0 |
10 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:3-hydroxyanthranilic acid dioxygenase-like |
| 88 | c3ctrA_ |
|
not modelled |
7.0 |
57 |
PDB header:hydrolase
Chain: A: PDB Molecule:poly(a)-specific ribonuclease parn;
PDBTitle: crystal structure of the rrm-domain of the poly(a)-specific2 ribonuclease parn bound to m7gtp
|
| 89 | d2d8za1 |
|
not modelled |
6.9 |
43 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 90 | d1sq4a_ |
|
not modelled |
6.9 |
14 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:YlbA-like |
| 91 | c3rnsA_ |
|
not modelled |
6.5 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:cupin 2 conserved barrel domain protein;
PDBTitle: cupin 2 conserved barrel domain protein from leptotrichia buccalis
|
| 92 | d1txka1 |
|
not modelled |
6.4 |
24 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:E-set domains of sugar-utilizing enzymes |
| 93 | c3bu7A_ |
|
not modelled |
6.4 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:gentisate 1,2-dioxygenase;
PDBTitle: crystal structure and biochemical characterization of gdosp,2 a gentisate 1,2-dioxygenase from silicibacter pomeroyi
|
| 94 | d3bu7a1 |
|
not modelled |
6.4 |
19 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Gentisate 1,2-dioxygenase-like |
| 95 | d2cura2 |
|
not modelled |
6.0 |
36 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 96 | d3ehwa1 |
|
not modelled |
6.0 |
31 |
Fold:beta-clip
Superfamily:dUTPase-like
Family:dUTPase-like |
| 97 | c2pfwB_ |
|
not modelled |
6.0 |
14 |
PDB header:unknown function
Chain: B: PDB Molecule:cupin 2, conserved barrel domain protein;
PDBTitle: crystal structure of a rmlc-like cupin (sfri_3105) from shewanella2 frigidimarina ncimb 400 at 1.90 a resolution
|
| 98 | d1ei5a2 |
|
not modelled |
5.9 |
13 |
Fold:Streptavidin-like
Superfamily:D-aminopeptidase, middle and C-terminal domains
Family:D-aminopeptidase, middle and C-terminal domains |
| 99 | d1zrra1 |
|
not modelled |
5.7 |
11 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Acireductone dioxygenase |