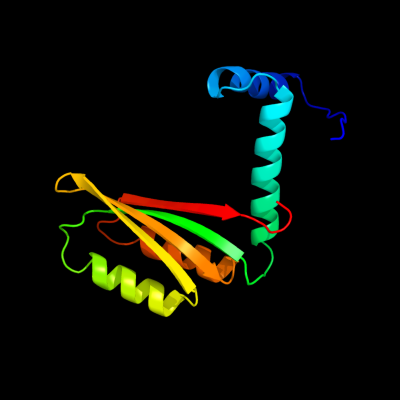
| 1 | c2ca9B_
|
|
 |
100.0 |
29 |
PDB header:transcriptional regulation
Chain: B: PDB Molecule:putative nickel-responsive regulator;
PDBTitle: apo-nikr from helicobacter pylori in closed trans-2 conformation
|
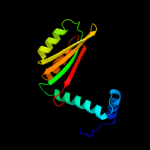
| 2 | c2bj3D_
|
|
 |
100.0 |
32 |
PDB header:transcription
Chain: D: PDB Molecule:nickel responsive regulator;
PDBTitle: nikr-apo
|
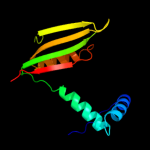
| 3 | c1q5vB_
|
|
 |
100.0 |
100 |
PDB header:transcription
Chain: B: PDB Molecule:nickel responsive regulator;
PDBTitle: apo-nikr
|
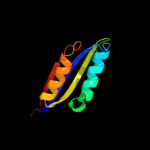
| 4 | d1q5ya_
|
|
 |
100.0 |
100 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Nickel responsive regulator NikR, C-terminal domain |
| 5 | d2bj7a2
|
|
 |
100.0 |
29 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Nickel responsive regulator NikR, C-terminal domain |
| 6 | c2y3yC_
|
|
 |
100.0 |
29 |
PDB header:transcription
Chain: C: PDB Molecule:putative nickel-responsive regulator;
PDBTitle: holo-ni(ii) hpnikr is a symmetric tetramer containing four2 canonic square-planar ni(ii) ions at physiological ph
|
| 7 | d2bj7a1
|
|
 |
99.6 |
35 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
| 8 | d2hzaa1
|
|
 |
99.6 |
98 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
| 9 | d2hzab1
|
|
 |
99.5 |
98 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
| 10 | c2k5jB_
|
|
 |
97.8 |
20 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein yiif;
PDBTitle: solution structure of protein yiif from shigella flexneri2 serotype 5b (strain 8401) . northeast structural genomics3 consortium target sft1
|
| 11 | c3kxeD_
|
|
 |
97.3 |
31 |
PDB header:protein binding
Chain: D: PDB Molecule:antitoxin protein pard-1;
PDBTitle: a conserved mode of protein recognition and binding in a2 pard-pare toxin-antitoxin complex
|
| 12 | d2nzca1
|
|
 |
97.0 |
17 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:TM1266-like |
| 13 | d2cpga_
|
|
 |
93.2 |
32 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
| 14 | c2k29A_
|
|
 |
91.5 |
26 |
PDB header:transcription
Chain: A: PDB Molecule:antitoxin relb;
PDBTitle: structure of the dbd domain of e. coli antitoxin relb
|
| 15 | c2kelB_
|
|
 |
90.2 |
20 |
PDB header:transcription repressor
Chain: B: PDB Molecule:uncharacterized protein 56b;
PDBTitle: structure of the transcription regulator svtr from the2 hyperthermophilic archaeal virus sirv1
|
| 16 | c3h87D_
|
|
 |
77.8 |
44 |
PDB header:toxin/antitoxin
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: rv0301 rv0300 toxin antitoxin complex from mycobacterium tuberculosis
|
| 17 | d1y9ba1
|
|
 |
64.2 |
16 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:VCA0319-like |
| 18 | c2k9iB_
|
|
 |
59.3 |
12 |
PDB header:dna binding protein
Chain: B: PDB Molecule:plasmid prn1, complete sequence;
PDBTitle: nmr structure of plasmid copy control protein orf56 from2 sulfolobus islandicus
|
| 19 | c3gxqB_
|
|
 |
55.8 |
31 |
PDB header:dna binding protein/dna
Chain: B: PDB Molecule:putative regulator of transfer genes arta;
PDBTitle: structure of arta and dna complex
|
| 20 | d1p94a_
|
|
 |
55.1 |
14 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
| 21 | d1ygya3 |
|
not modelled |
36.6 |
8 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain |
| 22 | d1vioa2 |
|
not modelled |
34.4 |
29 |
Fold:Alpha-L RNA-binding motif
Superfamily:Alpha-L RNA-binding motif
Family:Pseudouridine synthase RsuA N-terminal domain |
| 23 | c2rbfB_ |
|
not modelled |
32.3 |
26 |
PDB header:oxidoreductase/dna
Chain: B: PDB Molecule:bifunctional protein puta;
PDBTitle: structure of the ribbon-helix-helix domain of escherichia coli puta2 (puta52) complexed with operator dna (o2)
|
| 24 | d2cyya2 |
|
not modelled |
31.9 |
15 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Lrp/AsnC-like transcriptional regulator C-terminal domain |
| 25 | c2knjA_ |
|
not modelled |
31.7 |
21 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:microplusin preprotein;
PDBTitle: nmr structure of microplusin a antimicrobial peptide from2 rhipicephalus (boophilus) microplus
|
| 26 | c2e1cA_ |
|
not modelled |
25.9 |
15 |
PDB header:transcription/dna
Chain: A: PDB Molecule:putative hth-type transcriptional regulator ph1519;
PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
|
| 27 | d1c06a_ |
|
not modelled |
25.2 |
36 |
Fold:Alpha-L RNA-binding motif
Superfamily:Alpha-L RNA-binding motif
Family:Ribosomal protein S4 |
| 28 | d1sc6a3 |
|
not modelled |
24.9 |
7 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain |
| 29 | c2vixB_ |
|
not modelled |
24.5 |
11 |
PDB header:transport protein
Chain: B: PDB Molecule:protein mxic;
PDBTitle: methylated shigella flexneri mxic
|
| 30 | d1l3la1 |
|
not modelled |
24.3 |
10 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:GerE-like (LuxR/UhpA family of transcriptional regulators) |
| 31 | d1p9ka_ |
|
not modelled |
23.6 |
22 |
Fold:Alpha-L RNA-binding motif
Superfamily:Alpha-L RNA-binding motif
Family:YbcJ-like |
| 32 | c2exuA_ |
|
not modelled |
23.0 |
3 |
PDB header:transcription
Chain: A: PDB Molecule:transcription initiation protein spt4/spt5;
PDBTitle: crystal structure of saccharomyces cerevisiae transcription elongation2 factors spt4-spt5ngn domain
|
| 33 | c2q2kA_ |
|
not modelled |
21.1 |
36 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: structure of nucleic-acid binding protein
|
| 34 | d1u8sa1 |
|
not modelled |
20.7 |
3 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Glycine cleavage system transcriptional repressor |
| 35 | c3m8jA_ |
|
not modelled |
20.2 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:focb protein;
PDBTitle: crystal structure of e.coli focb at 1.4 a resolution
|
| 36 | c2qhoF_ |
|
not modelled |
19.2 |
27 |
PDB header:protein binding/ligase
Chain: F: PDB Molecule:e3 ubiquitin-protein ligase edd1;
PDBTitle: crystal structure of the uba domain from edd ubiquitin2 ligase in complex with ubiquitin
|
| 37 | c2kpoA_ |
|
not modelled |
17.3 |
22 |
PDB header:de novo protein
Chain: A: PDB Molecule:rossmann 2x2 fold protein;
PDBTitle: solution nmr structure of de novo designed rossmann 2x2 fold protein,2 northeast structural genomics consortium target or16
|
| 38 | c1u8sB_ |
|
not modelled |
16.6 |
5 |
PDB header:transcription
Chain: B: PDB Molecule:glycine cleavage system transcriptional
PDBTitle: crystal structure of putative glycine cleavage system2 transcriptional repressor
|
| 39 | c2cqjA_ |
|
not modelled |
16.2 |
13 |
PDB header:rna binding protein
Chain: A: PDB Molecule:u3 small nucleolar ribonucleoprotein protein
PDBTitle: solution structure of the s4 domain of u3 small nucleolar2 ribonucleoprotein protein imp3 homolog
|
| 40 | c2nyiB_ |
|
not modelled |
16.1 |
5 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:unknown protein;
PDBTitle: crystal structure of an unknown protein from galdieria2 sulphuraria
|
| 41 | c2rnjA_ |
|
not modelled |
15.7 |
9 |
PDB header:transcription
Chain: A: PDB Molecule:response regulator protein vrar;
PDBTitle: nmr structure of the s. aureus vrar dna binding domain
|
| 42 | c2k6lA_ |
|
not modelled |
15.5 |
23 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: the solution structure of xacb0070 from xanthonomas2 axonopodis pv citri reveals this new protein is a member of3 the rhh family of transcriptional repressors
|
| 43 | d2gnoa1 |
|
not modelled |
15.3 |
11 |
Fold:post-AAA+ oligomerization domain-like
Superfamily:post-AAA+ oligomerization domain-like
Family:DNA polymerase III clamp loader subunits, C-terminal domain |
| 44 | d1vhwa_ |
|
not modelled |
15.0 |
22 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 45 | d1xdia2 |
|
not modelled |
14.5 |
25 |
Fold:CO dehydrogenase flavoprotein C-domain-like
Superfamily:FAD/NAD-linked reductases, dimerisation (C-terminal) domain
Family:FAD/NAD-linked reductases, dimerisation (C-terminal) domain |
| 46 | d2ac7a1 |
|
not modelled |
14.5 |
19 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 47 | d1f06a2 |
|
not modelled |
14.5 |
32 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:Dihydrodipicolinate reductase-like |
| 48 | c2x3dC_ |
|
not modelled |
12.6 |
18 |
PDB header:unknown function
Chain: C: PDB Molecule:sso6206;
PDBTitle: crystal structure of sso6206 from sulfolobus solfataricus p2
|
| 49 | d1k9sa_ |
|
not modelled |
12.5 |
22 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 50 | c2krfB_ |
|
not modelled |
12.5 |
16 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulatory protein coma;
PDBTitle: nmr solution structure of the dna binding domain of competence protein2 a
|
| 51 | c3kk4B_ |
|
not modelled |
12.4 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein bp1543;
PDBTitle: uncharacterized protein bp1543 from bordetella pertussis tohama i
|
| 52 | c3mntA_ |
|
not modelled |
11.8 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:leucine carboxyl methyltransferase 1;
PDBTitle: crystal structure of human leucine carboxyl methyltransferase 1
|
| 53 | c2y9jt_ |
|
not modelled |
11.8 |
12 |
PDB header:protein transport
Chain: T: PDB Molecule:protein prgh;
PDBTitle: three-dimensional model of salmonella's needle complex at2 subnanometer resolution
|
| 54 | c2jpcA_ |
|
not modelled |
11.5 |
7 |
PDB header:dna binding protein
Chain: A: PDB Molecule:ssrb;
PDBTitle: ssrb dna binding protein
|
| 55 | c1zljE_ |
|
not modelled |
11.3 |
12 |
PDB header:transcription
Chain: E: PDB Molecule:dormancy survival regulator;
PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic2 response regulator dosr c-terminal domain
|
| 56 | d1sxjd1 |
|
not modelled |
10.9 |
10 |
Fold:post-AAA+ oligomerization domain-like
Superfamily:post-AAA+ oligomerization domain-like
Family:DNA polymerase III clamp loader subunits, C-terminal domain |
| 57 | c3b47A_ |
|
not modelled |
10.5 |
23 |
PDB header:signaling protein
Chain: A: PDB Molecule:methyl-accepting chemotaxis protein;
PDBTitle: periplasmic sensor domain of chemotaxis protein gsu0582
|
| 58 | d2nwua1 |
|
not modelled |
10.5 |
30 |
Fold:RL5-like
Superfamily:RL5-like
Family:SSO1042-like |
| 59 | c1yj7A_ |
|
not modelled |
9.9 |
14 |
PDB header:protein transport
Chain: A: PDB Molecule:escj;
PDBTitle: crystal structure of enteropathogenic e.coli (epec) type iii secretion2 system protein escj
|
| 60 | d1uyla_ |
|
not modelled |
9.7 |
17 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:Heat shock protein 90, HSP90, N-terminal domain |
| 61 | c2fwyA_ |
|
not modelled |
9.7 |
17 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein hsp 90-alpha;
PDBTitle: structure of human hsp90-alpha bound to the potent water2 soluble inhibitor pu-h64
|
| 62 | c1x3uA_ |
|
not modelled |
9.6 |
6 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulatory protein fixj;
PDBTitle: solution structure of the c-terminal transcriptional2 activator domain of fixj from sinorhizobium melilot
|
| 63 | c3ieiD_ |
|
not modelled |
9.4 |
21 |
PDB header:transferase
Chain: D: PDB Molecule:leucine carboxyl methyltransferase 1;
PDBTitle: crystal structure of human leucine carboxylmethyltransferase-1 in2 complex with s-adenosyl homocysteine
|
| 64 | d2qy9a1 |
|
not modelled |
9.4 |
16 |
Fold:Four-helical up-and-down bundle
Superfamily:Domain of the SRP/SRP receptor G-proteins
Family:Domain of the SRP/SRP receptor G-proteins |
| 65 | d1sr9a1 |
|
not modelled |
9.3 |
17 |
Fold:RuvA C-terminal domain-like
Superfamily:post-HMGL domain-like
Family:DmpG/LeuA communication domain-like |
| 66 | d2raqa1 |
|
not modelled |
9.3 |
18 |
Fold:Ferredoxin-like
Superfamily:MTH889-like
Family:MTH889-like |
| 67 | c2hwyB_ |
|
not modelled |
9.2 |
25 |
PDB header:rna binding protein
Chain: B: PDB Molecule:protein smg5;
PDBTitle: structure of pin domain of human smg5.
|
| 68 | c2nysA_ |
|
not modelled |
9.2 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:agr_c_3712p;
PDBTitle: x-ray crystal structure of protein agr_c_3712 from2 agrobacterium tumefaciens. northeast structural genomics3 consortium target atr88.
|
| 69 | d2nysa1 |
|
not modelled |
9.2 |
18 |
Fold:SspB-like
Superfamily:SspB-like
Family:AGR C 3712p-like |
| 70 | c2kebA_ |
|
not modelled |
9.2 |
19 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna polymerase subunit alpha b;
PDBTitle: nmr solution structure of the n-terminal domain of the dna polymerase2 alpha p68 subunit
|
| 71 | c3dh3C_ |
|
not modelled |
8.8 |
31 |
PDB header:isomerase/rna
Chain: C: PDB Molecule:ribosomal large subunit pseudouridine synthase f;
PDBTitle: crystal structure of rluf in complex with a 22 nucleotide2 rna substrate
|
| 72 | d1rjda_ |
|
not modelled |
8.8 |
18 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Leucine carboxy methyltransferase Ppm1 |
| 73 | d2cg4a2 |
|
not modelled |
8.2 |
10 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Lrp/AsnC-like transcriptional regulator C-terminal domain |
| 74 | c3ggzH_ |
|
not modelled |
8.1 |
36 |
PDB header:protein transport, endocytosis
Chain: H: PDB Molecule:vacuolar protein-sorting-associated protein 46;
PDBTitle: crystal structure of s.cerevisiae ist1 n-terminal domain in2 complex with did2 mim motif
|
| 75 | c2e1aD_ |
|
not modelled |
8.1 |
10 |
PDB header:transcription
Chain: D: PDB Molecule:75aa long hypothetical regulatory protein asnc;
PDBTitle: crystal structure of ffrp-dm1
|
| 76 | d1ebda3 |
|
not modelled |
8.1 |
19 |
Fold:CO dehydrogenase flavoprotein C-domain-like
Superfamily:FAD/NAD-linked reductases, dimerisation (C-terminal) domain
Family:FAD/NAD-linked reductases, dimerisation (C-terminal) domain |
| 77 | c3b42B_ |
|
not modelled |
8.0 |
22 |
PDB header:signaling protein
Chain: B: PDB Molecule:methyl-accepting chemotaxis protein, putative;
PDBTitle: periplasmic sensor domain of chemotaxis protein gsu0935
|
| 78 | d1lvla3 |
|
not modelled |
8.0 |
25 |
Fold:CO dehydrogenase flavoprotein C-domain-like
Superfamily:FAD/NAD-linked reductases, dimerisation (C-terminal) domain
Family:FAD/NAD-linked reductases, dimerisation (C-terminal) domain |
| 79 | c3ggzF_ |
|
not modelled |
8.0 |
36 |
PDB header:protein transport, endocytosis
Chain: F: PDB Molecule:vacuolar protein-sorting-associated protein 46;
PDBTitle: crystal structure of s.cerevisiae ist1 n-terminal domain in2 complex with did2 mim motif
|
| 80 | c3ggzE_ |
|
not modelled |
8.0 |
36 |
PDB header:protein transport, endocytosis
Chain: E: PDB Molecule:vacuolar protein-sorting-associated protein 46;
PDBTitle: crystal structure of s.cerevisiae ist1 n-terminal domain in2 complex with did2 mim motif
|
| 81 | c3ggzG_ |
|
not modelled |
7.9 |
36 |
PDB header:protein transport, endocytosis
Chain: G: PDB Molecule:vacuolar protein-sorting-associated protein 46;
PDBTitle: crystal structure of s.cerevisiae ist1 n-terminal domain in2 complex with did2 mim motif
|
| 82 | d1mnta_ |
|
not modelled |
7.8 |
14 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:Arc/Mnt-like phage repressors |
| 83 | d1qxpa2 |
|
not modelled |
7.8 |
9 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Penta-EF-hand proteins |
| 84 | d3bpda1 |
|
not modelled |
7.8 |
18 |
Fold:Ferredoxin-like
Superfamily:MTH889-like
Family:MTH889-like |
| 85 | c2ia0A_ |
|
not modelled |
7.6 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative hth-type transcriptional regulator pf0864;
PDBTitle: transcriptional regulatory protein pf0864 from pyrococcus furiosus a2 member of the asnc family (pf0864)
|
| 86 | c2c45F_ |
|
not modelled |
7.5 |
36 |
PDB header:lyase
Chain: F: PDB Molecule:aspartate 1-decarboxylase precursor;
PDBTitle: native precursor of pyruvoyl dependent aspartate2 decarboxylase
|
| 87 | d1zpva1 |
|
not modelled |
7.4 |
8 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:SP0238-like |
| 88 | d1cjya2 |
|
not modelled |
7.3 |
13 |
Fold:FabD/lysophospholipase-like
Superfamily:FabD/lysophospholipase-like
Family:Lysophospholipase |
| 89 | d1nm2a2 |
|
not modelled |
7.3 |
16 |
Fold:Ferredoxin-like
Superfamily:Probable ACP-binding domain of malonyl-CoA ACP transacylase
Family:Probable ACP-binding domain of malonyl-CoA ACP transacylase |
| 90 | d1odka_ |
|
not modelled |
7.2 |
28 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 91 | d2pzza1 |
|
not modelled |
7.2 |
21 |
Fold:RL5-like
Superfamily:RL5-like
Family:SSO1042-like |
| 92 | c3m6jD_ |
|
not modelled |
7.2 |
28 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of unknown function protein from leptospirillum2 rubarum
|
| 93 | c3h7hB_ |
|
not modelled |
7.2 |
17 |
PDB header:transcription
Chain: B: PDB Molecule:transcription elongation factor spt5;
PDBTitle: crystal structure of the human transcription elongation factor dsif,2 hspt4/hspt5 (176-273)
|
| 94 | c2zwaA_ |
|
not modelled |
7.1 |
48 |
PDB header:transferase
Chain: A: PDB Molecule:leucine carboxyl methyltransferase 2;
PDBTitle: crystal structure of trna wybutosine synthesizing enzyme2 tyw4
|
| 95 | c2qasA_ |
|
not modelled |
7.1 |
16 |
PDB header:hydrolase activator
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of caulobacter crescentus sspb ortholog
|
| 96 | c2qazC_ |
|
not modelled |
7.0 |
16 |
PDB header:hydrolase activator
Chain: C: PDB Molecule:sspb protein;
PDBTitle: structure of c. crescentus sspb ortholog
|
| 97 | c3qv0A_ |
|
not modelled |
7.0 |
38 |
PDB header:protein binding
Chain: A: PDB Molecule:mitochondrial acidic protein mam33;
PDBTitle: crystal structure of saccharomyces cerevisiae mam33
|
| 98 | d1zrra1 |
|
not modelled |
6.9 |
21 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Acireductone dioxygenase |
| 99 | d2hs5a1 |
|
not modelled |
6.9 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:GntR-like transcriptional regulators |