| 1 |
|
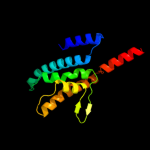
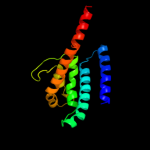
PDB 3k07 chain A
Region: 8 - 169
Aligned: 137
Modelled: 137
Confidence: 48.0%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:cation efflux system protein cusa;
PDBTitle: crystal structure of cusa
Phyre2
| 2 |
|
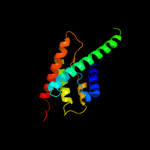
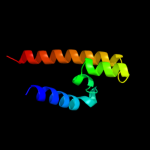
PDB 1oy8 chain A
Region: 13 - 169
Aligned: 132
Modelled: 132
Confidence: 38.3%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:acriflavine resistance protein b;
PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
Phyre2
| 3 |
|
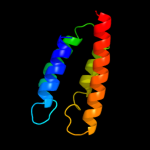
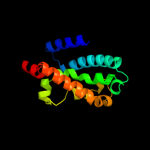
PDB 1j4n chain A
Region: 351 - 515
Aligned: 165
Modelled: 165
Confidence: 32.8%
Identity: 17%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2
| 4 |
|
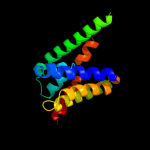
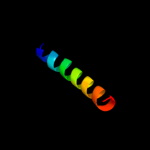
PDB 2r6g chain F domain 1
Region: 408 - 472
Aligned: 65
Modelled: 65
Confidence: 14.2%
Identity: 14%
Fold: MalF N-terminal region-like
Superfamily: MalF N-terminal region-like
Family: MalF N-terminal region-like
Phyre2
| 5 |
|
PDB 2ht2 chain B
Region: 17 - 182
Aligned: 119
Modelled: 138
Confidence: 14.0%
Identity: 15%
PDB header:membrane protein
Chain: B: PDB Molecule:h(+)/cl(-) exchange transporter clca;
PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
Phyre2
| 6 |
|
PDB 3k3g chain A
Region: 81 - 169
Aligned: 89
Modelled: 89
Confidence: 12.9%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:urea transporter;
PDBTitle: crystal structure of the urea transporter from desulfovibrio vulgaris2 bound to 1,3-dimethylurea
Phyre2
| 7 |
|
PDB 1kpl chain A
Region: 17 - 186
Aligned: 123
Modelled: 142
Confidence: 11.8%
Identity: 14%
Fold: Clc chloride channel
Superfamily: Clc chloride channel
Family: Clc chloride channel
Phyre2
| 8 |
|
PDB 1rc2 chain A
Region: 358 - 508
Aligned: 148
Modelled: 148
Confidence: 10.1%
Identity: 14%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2
| 9 |
|
PDB 2ksf chain A
Region: 411 - 502
Aligned: 91
Modelled: 92
Confidence: 8.6%
Identity: 11%
PDB header:transferase
Chain: A: PDB Molecule:sensor protein kdpd;
PDBTitle: backbone structure of the membrane domain of e. coli2 histidine kinase receptor kdpd, center for structures of3 membrane proteins (csmp) target 4312c
Phyre2
| 10 |
|
PDB 3aqp chain B
Region: 5 - 169
Aligned: 136
Modelled: 139
Confidence: 7.5%
Identity: 18%
PDB header:membrane protein
Chain: B: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
Phyre2
| 11 |
|
PDB 3llq chain B
Region: 356 - 503
Aligned: 145
Modelled: 148
Confidence: 7.4%
Identity: 19%
PDB header:membrane protein
Chain: B: PDB Molecule:aquaporin z 2;
PDBTitle: aquaporin structure from plant pathogen agrobacterium tumerfaciens
Phyre2
| 12 |
|
PDB 1ots chain A
Region: 11 - 186
Aligned: 129
Modelled: 148
Confidence: 7.2%
Identity: 15%
Fold: Clc chloride channel
Superfamily: Clc chloride channel
Family: Clc chloride channel
Phyre2
| 13 |
|
PDB 3gd8 chain A
Region: 358 - 427
Aligned: 67
Modelled: 70
Confidence: 6.5%
Identity: 10%
PDB header:membrane protein
Chain: A: PDB Molecule:aquaporin-4;
PDBTitle: crystal structure of human aquaporin 4 at 1.8 and its mechanism of2 conductance
Phyre2
| 14 |
|
PDB 3nd0 chain A
Region: 17 - 180
Aligned: 117
Modelled: 136
Confidence: 6.5%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:sll0855 protein;
PDBTitle: x-ray crystal structure of a slow cyanobacterial cl-/h+ antiporter
Phyre2
| 15 |
|
PDB 2e74 chain D domain 2
Region: 401 - 424
Aligned: 24
Modelled: 24
Confidence: 6.2%
Identity: 38%
Fold: Single transmembrane helix
Superfamily: ISP transmembrane anchor
Family: ISP transmembrane anchor
Phyre2
| 16 |
|
PDB 1iwg chain A domain 8
Region: 15 - 169
Aligned: 130
Modelled: 130
Confidence: 6.2%
Identity: 12%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 17 |
|
PDB 1s7b chain A
Region: 93 - 173
Aligned: 78
Modelled: 81
Confidence: 5.6%
Identity: 10%
Fold: Multidrug resistance efflux transporter EmrE
Superfamily: Multidrug resistance efflux transporter EmrE
Family: Multidrug resistance efflux transporter EmrE
Phyre2
| 18 |
|
PDB 2jp3 chain A
Region: 472 - 505
Aligned: 32
Modelled: 34
Confidence: 5.5%
Identity: 25%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 19 |
|
PDB 2jo1 chain A
Region: 472 - 503
Aligned: 30
Modelled: 32
Confidence: 5.5%
Identity: 27%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2