1 c2yjlC_
86.4
13
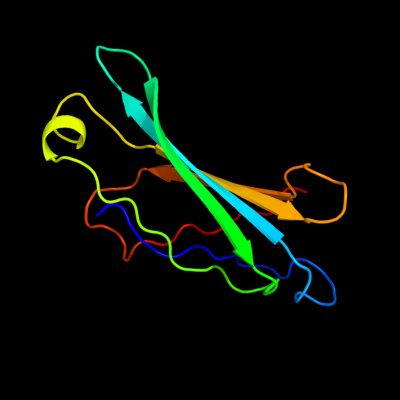
PDB header: lipid-binding proteinChain: C: PDB Molecule: exoenzyme s synthesis protein b;PDBTitle: structural characterization of a secretin pilot protein2 from the type iii secretion system (t3ss) of pseudomonas3 aeruginosa
2 d2b9va1
74.0
18
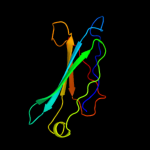
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: PepX C-terminal domain-like3 d1ju3a1
64.7
25
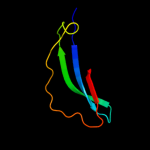
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: PepX C-terminal domain-like4 c1lnsA_
56.9
9
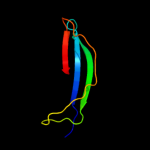
PDB header: hydrolaseChain: A: PDB Molecule: x-prolyl dipeptidyl aminopetidase;PDBTitle: crystal structure analysis of the x-prolyl dipeptidyl2 aminopeptidase from lactococcus lactis
5 c2b9vB_
52.6
21
PDB header: hydrolaseChain: B: PDB Molecule: alpha-amino acid ester hydrolase;PDBTitle: acetobacter turbidans alpha-amino acid ester hydrolase
6 c1mpxB_
51.9
25
PDB header: hydrolaseChain: B: PDB Molecule: alpha-amino acid ester hydrolase;PDBTitle: alpha-amino acid ester hydrolase labeled with selenomethionine
7 d1e5ra_
44.1
12
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: Type II Proline 3-hydroxylase (proline oxidase)8 c3cm1C_
43.0
20
PDB header: cell cycleChain: C: PDB Molecule: ssga-like sporulation-specific cell division protein;PDBTitle: crystal structure of ssga-like sporulation-specific cell division2 protein (yp_290167.1) from thermobifida fusca yx-er1 at 2.60 a3 resolution
9 c1l7qA_
40.6
23
PDB header: hydrolaseChain: A: PDB Molecule: cocaine esterase;PDBTitle: ser117ala mutant of bacterial cocaine esterase coce
10 d1tg7a2
38.3
8
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Beta-galactosidase LacA, domains 4 and 511 d2vnga1
37.7
14
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: NPCBM-like12 d1lnsa2
37.6
13
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: PepX C-terminal domain-like13 c3ib3A_
23.3
12
PDB header: hydrolaseChain: A: PDB Molecule: coce/nond family hydrolase;PDBTitle: crystal structure of sacol2612 - coce/nond family hydrolase from2 staphylococcus aureus
14 d2vmha1
21.1
12
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: NPCBM-like15 d1wioa2
19.8
26
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)16 c1ux6A_
16.5
13
PDB header: cell adhesionChain: A: PDB Molecule: thrombospondin-1;PDBTitle: structure of a thrombospondin c-terminal fragment reveals a2 novel calcium core in the type 3 repeats
17 d1mpxa1
15.4
25
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: PepX C-terminal domain-like18 c1wruA_
14.7
19
PDB header: structural proteinChain: A: PDB Molecule: 43 kda tail protein;PDBTitle: structure of central hub elucidated by x-ray analysis of gene product2 44; baseplate component of bacteriophage mu
19 d2ccla1
13.6
23
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Carbohydrate-binding domainFamily: Cellulose-binding domain family III20 d2icya1
12.7
29
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: GlmU C-terminal domain-like21 c2qeaB_
not modelled
12.7
13
PDB header: oxidoreductaseChain: B: PDB Molecule: putative general stress protein 26;PDBTitle: crystal structure of a putative general stress protein 26 (jann_0955)2 from jannaschia sp. ccs1 at 2.46 a resolution
22 c1yo8A_
not modelled
12.4
6
PDB header: cell adhesionChain: A: PDB Molecule: thrombospondin-2;PDBTitle: structure of the c-terminal domain of human thrombospondin-2
23 c2giaB_
not modelled
11.2
33
PDB header: translationChain: B: PDB Molecule: mitochondrial rna-binding protein 1;PDBTitle: crystal structures of trypanosoma bruciei mrp1/mrp2
24 d2giab1
not modelled
11.2
33
Fold: ssDNA-binding transcriptional regulator domainSuperfamily: ssDNA-binding transcriptional regulator domainFamily: Guide RNA binding protein gBP25 d1rhoa_
not modelled
11.1
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: RhoGDI-like26 d1yq2a3
not modelled
10.2
16
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain27 c2qv8B_
not modelled
10.2
22
PDB header: transport proteinChain: B: PDB Molecule: general secretion pathway protein h;PDBTitle: structure of the minor pseudopilin epsh from the type 2 secretion2 system of vibrio cholerae
28 d2oz4a1
not modelled
9.8
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: C2 set domains29 d1qhqa_
not modelled
9.1
9
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like30 d2vo8a1
not modelled
8.6
8
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Carbohydrate-binding domainFamily: Cellulose-binding domain family III31 c3hwjA_
not modelled
8.5
7
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase mycbp2;PDBTitle: crystal structure of the second phr domain of mouse myc-2 binding protein 2 (mycbp-2)
32 d2a6va1
not modelled
8.1
20
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Lectin leg-like33 c2ov7C_
not modelled
8.1
16
PDB header: ribosomal proteinChain: C: PDB Molecule: 50s ribosomal protein l1;PDBTitle: the first domain of the ribosomal protein l1 from thermus2 thermophilus
34 d1cida1
not modelled
8.1
28
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)35 c1cidA_
not modelled
7.7
32
PDB header: t-cell surface glycoproteinChain: A: PDB Molecule: t cell surface glycoprotein cd4;PDBTitle: crystal structure of domains 3 & 4 of rat cd4 and their2 relationship to the nh2-terminal domains
36 c1c8uA_
not modelled
7.3
14
PDB header: hydrolaseChain: A: PDB Molecule: acyl-coa thioesterase ii;PDBTitle: crystal structure of the e.coli thioesterase ii, a2 homologue of the human nef-binding enzyme
37 d1c1za5
not modelled
7.1
17
Fold: Complement control module/SCR domainSuperfamily: Complement control module/SCR domainFamily: Complement control module/SCR domain38 d1mnga1
not modelled
6.9
27
Fold: Long alpha-hairpinSuperfamily: Fe,Mn superoxide dismutase (SOD), N-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), N-terminal domain39 c3r7gB_
not modelled
6.8
18
PDB header: protein bindingChain: B: PDB Molecule: formin-2;PDBTitle: crystal structure of spire kind domain in complex with the tail of2 fmn2
40 c3fbyC_
not modelled
6.4
17
PDB header: cell adhesionChain: C: PDB Molecule: cartilage oligomeric matrix protein;PDBTitle: the crystal structure of the signature domain of cartilage oligomeric2 matrix protein.
41 d1ux6a1
not modelled
6.1
11
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Thrombospondin C-terminal domain42 d1aoha_
not modelled
5.8
31
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Carbohydrate-binding domainFamily: Cellulose-binding domain family III43 d2bs2c1
not modelled
5.5
19
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Fumarate reductase respiratory complex cytochrome b subunit, FrdC44 c3bbjA_
not modelled
5.2
0
PDB header: hydrolaseChain: A: PDB Molecule: putative thioesterase ii;PDBTitle: crystal structure of a putative thioesterase ii (tfu_2367) from2 thermobifida fusca yx at 2.45 a resolution
45 d1quba5
not modelled
5.2
17
Fold: Complement control module/SCR domainSuperfamily: Complement control module/SCR domainFamily: Complement control module/SCR domain46 d2apsa_
not modelled
5.1
20
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like