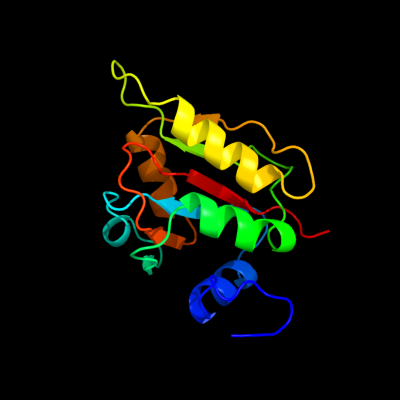
1 c2wcvI_
100.0
100
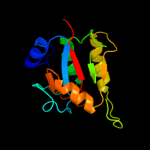
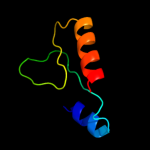
PDB header: isomeraseChain: I: PDB Molecule: l-fucose mutarotase;PDBTitle: crystal structure of bacterial fucu
2 d2ob5a1
100.0
33
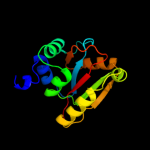
Fold: RbsD-likeSuperfamily: RbsD-likeFamily: RbsD-like3 c2wcuB_
100.0
47
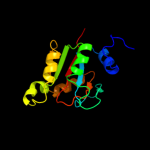
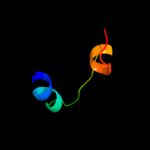
PDB header: isomeraseChain: B: PDB Molecule: protein fucu homolog;PDBTitle: crystal structure of mammalian fucu
4 c3mvkA_
100.0
41
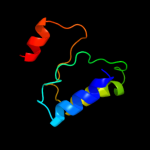
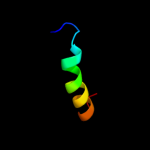
PDB header: isomeraseChain: A: PDB Molecule: protein fucu;PDBTitle: the crystal structure of fucu from bifidobacterium longum to 1.65a
5 c3e7nB_
100.0
18
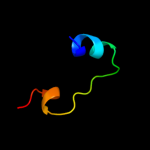
PDB header: transport proteinChain: B: PDB Molecule: d-ribose high-affinity transport system;PDBTitle: crystal structure of d-ribose high-affinity transport system from2 salmonella typhimurium lt2
6 d1ogda_
100.0
21
Fold: RbsD-likeSuperfamily: RbsD-likeFamily: RbsD-like7 c3p13B_
100.0
16
PDB header: isomeraseChain: B: PDB Molecule: d-ribose pyranase;PDBTitle: complex structure of d-ribose pyranase sa240 with d-ribose
8 d1qkia1
26.7
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain9 c1e1cA_
22.2
20
PDB header: isomeraseChain: A: PDB Molecule: methylmalonyl-coa mutase alpha chain;PDBTitle: methylmalonyl-coa mutase h244a mutant
10 d1e3da_
18.9
36
Fold: HydA/Nqo6-likeSuperfamily: HydA/Nqo6-likeFamily: Nickel-iron hydrogenase, small subunit11 c2dgbA_
17.6
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein purs;PDBTitle: structure of thermus thermophilus purs in the p21 form
12 d1t4aa_
16.3
18
Fold: PurS-likeSuperfamily: PurS-likeFamily: PurS subunit of FGAM synthetase13 d1frfs_
15.4
28
Fold: HydA/Nqo6-likeSuperfamily: HydA/Nqo6-likeFamily: Nickel-iron hydrogenase, small subunit14 d1cc1s_
15.1
32
Fold: HydA/Nqo6-likeSuperfamily: HydA/Nqo6-likeFamily: Nickel-iron hydrogenase, small subunit15 c2yx5A_
14.4
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0062 protein mj1593;PDBTitle: crystal structure of methanocaldococcus jannaschii purs, one of the2 subunits of formylglycinamide ribonucleotide amidotransferase in the3 purine biosynthetic pathway
16 c3myrE_
14.0
24
PDB header: oxidoreductaseChain: E: PDB Molecule: hydrogenase (nife) small subunit hyda;PDBTitle: crystal structure of [nife] hydrogenase from allochromatium vinosum in2 its ni-a state
17 d1wuis1
13.8
24
Fold: HydA/Nqo6-likeSuperfamily: HydA/Nqo6-likeFamily: Nickel-iron hydrogenase, small subunit18 c1h2aS_
13.7
24
PDB header: oxidoreductaseChain: S: PDB Molecule: hydrogenase;PDBTitle: single crystals of hydrogenase from desulfovibrio vulgaris
19 c3iymA_
13.0
24
PDB header: virusChain: A: PDB Molecule: capsid protein;PDBTitle: backbone trace of the capsid protein dimer of a fungal partitivirus2 from electron cryomicroscopy and homology modeling
20 d1guza1
11.4
25
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like21 d1gtda_
not modelled
10.8
35
Fold: PurS-likeSuperfamily: PurS-likeFamily: PurS subunit of FGAM synthetase22 d176la_
not modelled
10.7
27
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: Phage lysozyme23 c3rgwS_
not modelled
10.3
28
PDB header: oxidoreductase/oxidoreductaseChain: S: PDB Molecule: membrane-bound hydrogenase (nife) small subunit hoxk;PDBTitle: crystal structure at 1.5 a resolution of an h2-reduced, o2-tolerant2 hydrogenase from ralstonia eutropha unmasks a novel iron-sulfur3 cluster
24 c2wpnA_
not modelled
10.1
36
PDB header: oxidoreductaseChain: A: PDB Molecule: periplasmic [nifese] hydrogenase, small subunit;PDBTitle: structure of the oxidised, as-isolated nifese hydrogenase2 from d. vulgaris hildenborough
25 d5ldha1
not modelled
9.6
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like26 d1yq9a1
not modelled
8.8
35
Fold: HydA/Nqo6-likeSuperfamily: HydA/Nqo6-likeFamily: Nickel-iron hydrogenase, small subunit27 d1vs6z1
not modelled
8.0
36
Fold: L28p-likeSuperfamily: L28p-likeFamily: Ribosomal protein L31p28 d1llda1
not modelled
8.0
30
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like29 c1w1wF_
not modelled
7.7
20
PDB header: cell adhesionChain: F: PDB Molecule: sister chromatid cohesion protein 1;PDBTitle: sc smc1hd:scc1-c complex, atpgs
30 d1w1we_
not modelled
7.7
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Rad21/Rec8-like31 d1vpka2
not modelled
7.7
9
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase III, beta subunit32 d189la_
not modelled
7.6
27
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: Phage lysozyme33 d1ldna1
not modelled
6.9
35
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like34 d1p37a_
not modelled
6.9
27
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: Phage lysozyme35 d2ldxa1
not modelled
6.7
25
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like36 d1swya_
not modelled
6.4
27
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: Phage lysozyme37 d1yjma1
not modelled
6.2
23
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain38 d1gpma3
not modelled
5.9
30
Fold: Alpha-lytic protease prodomain-likeSuperfamily: GMP synthetase C-terminal dimerisation domainFamily: GMP synthetase C-terminal dimerisation domain39 d1hyha1
not modelled
5.9
30
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like40 d1vq3a_
not modelled
5.8
27
Fold: PurS-likeSuperfamily: PurS-likeFamily: PurS subunit of FGAM synthetase41 d1cp2a_
not modelled
5.8
26
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like42 c2b664_
not modelled
5.6
36
PDB header: ribosomeChain: 4: PDB Molecule: 50s ribosomal protein l31;PDBTitle: 50s ribosomal subunit from a crystal structure of release factor rf1,2 trnas and mrna bound to the ribosome. this file contains the 50s3 subunit from a crystal structure of release factor rf1, trnas and4 mrna bound to the ribosome and is described in remark 400
43 c2xd4A_
not modelled
5.6
9
PDB header: ligaseChain: A: PDB Molecule: phosphoribosylamine--glycine ligase;PDBTitle: nucleotide-bound structures of bacillus subtilis glycinamide2 ribonucleotide synthetase
44 d1jkea_
not modelled
5.5
25
Fold: DTD-likeSuperfamily: DTD-likeFamily: DTD-like45 c1yj5C_
not modelled
5.5
23
PDB header: transferaseChain: C: PDB Molecule: 5' polynucleotide kinase-3' phosphatase fha domain;PDBTitle: molecular architecture of mammalian polynucleotide kinase, a dna2 repair enzyme
46 d1llca1
not modelled
5.5
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like47 c2zw2B_
not modelled
5.4
18
PDB header: ligaseChain: B: PDB Molecule: putative uncharacterized protein sts178;PDBTitle: crystal structure of formylglycinamide ribonucleotide amidotransferase2 iii from sulfolobus tokodaii (stpurs)
48 c3hl4B_
not modelled
5.2
10
PDB header: transferaseChain: B: PDB Molecule: choline-phosphate cytidylyltransferase a;PDBTitle: crystal structure of a mammalian ctp:phosphocholine2 cytidylyltransferase with cdp-choline
49 c2j034_
not modelled
5.1
36
PDB header: ribosomeChain: 4: PDB Molecule: 50s ribosomal protein l31;PDBTitle: structure of the thermus thermophilus 70s ribosome2 complexed with mrna, trna and paromomycin (part 4 of 4).3 this file contains the 50s subunit from molecule ii.