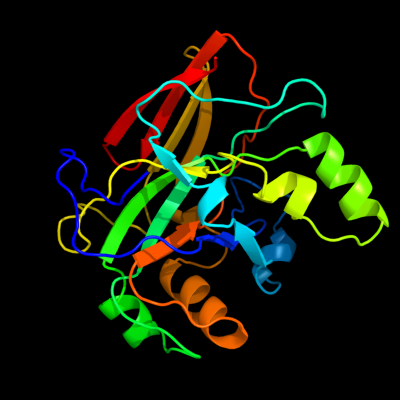
| 1 | d1nr9a_
|
|
 |
100.0 |
99 |
Fold:FAH
Superfamily:FAH
Family:FAH |
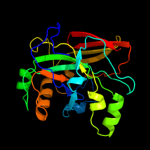
| 2 | c1i7oC_
|
|
 |
100.0 |
39 |
PDB header:isomerase, lyase
Chain: C: PDB Molecule:4-hydroxyphenylacetate degradation bifunctional
PDBTitle: crystal structure of hpce
|
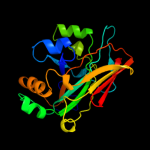
| 3 | c3l53F_
|
|
 |
100.0 |
46 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:putative fumarylacetoacetate isomerase/hydrolase;
PDBTitle: crystal structure of a putative fumarylacetoacetate2 isomerase/hydrolase from oleispira antarctica
|
| 4 | c3r6oA_
|
|
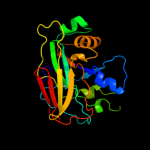 |
100.0 |
31 |
PDB header:isomerase
Chain: A: PDB Molecule:2-hydroxyhepta-2,4-diene-1, 7-dioateisomerase;
PDBTitle: crystal structure of a probable 2-hydroxyhepta-2,4-diene-1, 7-2 dioateisomerase from mycobacterium abscessus
|
| 5 | d1gtta2
|
|
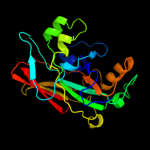 |
100.0 |
39 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 6 | c2dfuB_
|
|
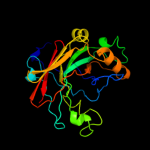 |
100.0 |
36 |
PDB header:isomerase
Chain: B: PDB Molecule:probable 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase;
PDBTitle: crystal structure of the 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase2 from thermus thermophilus hb8
|
| 7 | c1wzoC_
|
|
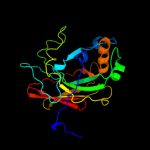 |
100.0 |
35 |
PDB header:isomerase
Chain: C: PDB Molecule:hpce;
PDBTitle: crystal structure of the hpce from thermus thermophilus hb8
|
| 8 | c3lzkC_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: C: PDB Molecule:fumarylacetoacetate hydrolase family protein;
PDBTitle: the crystal structure of a probably aromatic amino acid2 degradation protein from sinorhizobium meliloti 1021
|
| 9 | c3qdfA_
|
|
 |
100.0 |
38 |
PDB header:isomerase
Chain: A: PDB Molecule:2-hydroxyhepta-2,4-diene-1,7-dioate isomerase;
PDBTitle: crystal structure of 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase2 from mycobacterium marinum
|
| 10 | d1gtta1
|
|
 |
100.0 |
29 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 11 | d1sawa_
|
|
 |
100.0 |
47 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 12 | d1nkqa_
|
|
 |
100.0 |
36 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 13 | c2q1dX_
|
|
 |
100.0 |
22 |
PDB header:lyase
Chain: X: PDB Molecule:2-keto-3-deoxy-d-arabinonate dehydratase;
PDBTitle: 2-keto-3-deoxy-d-arabinonate dehydratase complexed with magnesium and2 2,5-dioxopentanoate
|
| 14 | d1hyoa2
|
|
 |
100.0 |
24 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 15 | c1hyoB_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:fumarylacetoacetate hydrolase;
PDBTitle: crystal structure of fumarylacetoacetate hydrolase2 complexed with 4-(hydroxymethylphosphinoyl)-3-oxo-butanoic3 acid
|
| 16 | d1sv6a_
|
|
 |
100.0 |
18 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 17 | c2eb5D_
|
|
 |
100.0 |
19 |
PDB header:lyase
Chain: D: PDB Molecule:2-oxo-hept-3-ene-1,7-dioate hydratase;
PDBTitle: crystal structure of hpcg complexed with oxalate
|
| 18 | d1c3ha_
|
|
 |
56.5 |
16 |
Fold:TNF-like
Superfamily:TNF-like
Family:TNF-like |
| 19 | d2cu3a1
|
|
 |
51.5 |
21 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:ThiS |
| 20 | c2kl0A_
|
|
 |
43.3 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative thiamin biosynthesis this;
PDBTitle: solution nmr structure of rhodopseudomonas palustris rpa3574,2 northeast structural genomics consortium (nesg) target rpr325
|
| 21 | c3cwiA_ |
|
not modelled |
36.7 |
18 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:thiamine-biosynthesis protein this;
PDBTitle: crystal structure of thiamine biosynthesis protein (this)2 from geobacter metallireducens. northeast structural3 genomics consortium target gmr137
|
| 22 | d1gr3a_ |
|
not modelled |
35.4 |
13 |
Fold:TNF-like
Superfamily:TNF-like
Family:TNF-like |
| 23 | c1gr3A_ |
|
not modelled |
35.4 |
13 |
PDB header:collagen
Chain: A: PDB Molecule:collagen x;
PDBTitle: structure of the human collagen x nc1 trimer
|
| 24 | d2evra2 |
|
not modelled |
34.9 |
37 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:NlpC/P60 |
| 25 | d1zud21 |
|
not modelled |
30.9 |
13 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:ThiS |
| 26 | d1pk6c_ |
|
not modelled |
29.4 |
21 |
Fold:TNF-like
Superfamily:TNF-like
Family:TNF-like |
| 27 | d2vbua1 |
|
not modelled |
29.2 |
31 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin kinase-like
Family:CTP-dependent riboflavin kinase-like |
| 28 | d1tygb_ |
|
not modelled |
28.9 |
16 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:MoaD/ThiS
Family:ThiS |
| 29 | c2xivA_ |
|
not modelled |
25.6 |
29 |
PDB header:structural protein
Chain: A: PDB Molecule:hypothetical invasion protein;
PDBTitle: structure of rv1477, hypothetical invasion protein of2 mycobacterium tuberculosis
|
| 30 | c1tygG_ |
|
not modelled |
25.3 |
16 |
PDB header:biosynthetic protein
Chain: G: PDB Molecule:yjbs;
PDBTitle: structure of the thiazole synthase/this complex
|
| 31 | d2f4la1 |
|
not modelled |
22.1 |
35 |
Fold:CUB-like
Superfamily:Acetamidase/Formamidase-like
Family:Acetamidase/Formamidase-like |
| 32 | d2g1la1 |
|
not modelled |
19.3 |
35 |
Fold:SMAD/FHA domain
Superfamily:SMAD/FHA domain
Family:FHA domain |
| 33 | d1t8sa_ |
|
not modelled |
19.2 |
12 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 34 | d1o91a_ |
|
not modelled |
18.3 |
20 |
Fold:TNF-like
Superfamily:TNF-like
Family:TNF-like |
| 35 | c1o91B_ |
|
not modelled |
18.3 |
20 |
PDB header:collagen
Chain: B: PDB Molecule:collagen alpha 1(viii) chain;
PDBTitle: crystal structure of a collagen viii nc1 domain trimer
|
| 36 | d1pk6a_ |
|
not modelled |
17.0 |
21 |
Fold:TNF-like
Superfamily:TNF-like
Family:TNF-like |
| 37 | c3gt2A_ |
|
not modelled |
15.2 |
30 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the p60 domain from m. avium2 paratuberculosis antigen map1272c
|
| 38 | c2ka3C_ |
|
not modelled |
13.9 |
32 |
PDB header:structural protein
Chain: C: PDB Molecule:emilin-1;
PDBTitle: structure of emilin-1 c1q-like domain
|
| 39 | c2fg0B_ |
|
not modelled |
13.1 |
39 |
PDB header:hydrolase
Chain: B: PDB Molecule:cog0791: cell wall-associated hydrolases (invasion-
PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
|
| 40 | d1pk6b_ |
|
not modelled |
12.6 |
20 |
Fold:TNF-like
Superfamily:TNF-like
Family:TNF-like |
| 41 | d1mvfd_ |
|
not modelled |
12.1 |
20 |
Fold:Double-split beta-barrel
Superfamily:AbrB/MazE/MraZ-like
Family:Kis/PemI addiction antidote |
| 42 | c2pkpA_ |
|
not modelled |
10.9 |
33 |
PDB header:lyase
Chain: A: PDB Molecule:homoaconitase small subunit;
PDBTitle: crystal structure of 3-isopropylmalate dehydratase (leud)2 from methhanocaldococcus jannaschii dsm2661 (mj1271)
|
| 43 | d1dm9a_ |
|
not modelled |
10.9 |
18 |
Fold:Alpha-L RNA-binding motif
Superfamily:Alpha-L RNA-binding motif
Family:Heat shock protein 15 kD |
| 44 | c1dm9A_ |
|
not modelled |
10.9 |
18 |
PDB header:structural genomics
Chain: A: PDB Molecule:hypothetical 15.5 kd protein in mrca-pcka
PDBTitle: heat shock protein 15 kd
|
| 45 | d2qamc2 |
|
not modelled |
9.8 |
46 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 46 | c3m9bK_ |
|
not modelled |
9.7 |
26 |
PDB header:chaperone
Chain: K: PDB Molecule:proteasome-associated atpase;
PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
|
| 47 | d1ds1a_ |
|
not modelled |
9.4 |
11 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Clavaminate synthase |
| 48 | c3g8qA_ |
|
not modelled |
9.2 |
13 |
PDB header:rna binding protein
Chain: A: PDB Molecule:predicted rna-binding protein, contains thump
PDBTitle: a cytidine deaminase edits c-to-u in transfer rnas in2 archaea
|
| 49 | c2e6zA_ |
|
not modelled |
8.7 |
32 |
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation factor spt5;
PDBTitle: solution structure of the second kow motif of human2 transcription elongation factor spt5
|
| 50 | d1rl2a2 |
|
not modelled |
8.4 |
31 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 51 | d1v7la_ |
|
not modelled |
8.1 |
49 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LeuD/IlvD-like
Family:LeuD-like |
| 52 | c3pbiA_ |
|
not modelled |
7.8 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:invasion protein;
PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
|
| 53 | d1x5na1 |
|
not modelled |
7.7 |
25 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 54 | d1g7sa1 |
|
not modelled |
7.6 |
20 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Translation proteins
Family:Elongation factors |
| 55 | c2z8uQ_ |
|
not modelled |
6.9 |
25 |
PDB header:transcription
Chain: Q: PDB Molecule:tata-box-binding protein;
PDBTitle: methanococcus jannaschii tbp
|
| 56 | c1gxcA_ |
|
not modelled |
6.9 |
21 |
PDB header:phosphoprotein-binding domain
Chain: A: PDB Molecule:serine/threonine-protein kinase chk2;
PDBTitle: fha domain from human chk2 kinase in complex with a2 synthetic phosphopeptide
|
| 57 | d1gxca_ |
|
not modelled |
6.9 |
21 |
Fold:SMAD/FHA domain
Superfamily:SMAD/FHA domain
Family:FHA domain |
| 58 | d2v4ja2 |
|
not modelled |
6.8 |
16 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:DsrA/DsrB N-terminal-domain-like |
| 59 | c3h1yA_ |
|
not modelled |
6.5 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:mannose-6-phosphate isomerase;
PDBTitle: crystal structure of mannose 6-phosphate isomerase from2 salmonella typhimurium bound to substrate (f6p)and metal3 atom (zn)
|
| 60 | c3mjjD_ |
|
not modelled |
6.5 |
31 |
PDB header:hydrolase
Chain: D: PDB Molecule:predicted acetamidase/formamidase;
PDBTitle: crystal structure analysis of a recombinant predicted2 acetamidase/formamidase from the thermophile thermoanaerobacter3 tengcongensis
|
| 61 | d2i4sa1 |
|
not modelled |
6.5 |
14 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:EpsC C-terminal domain-like |
| 62 | d2ba0a2 |
|
not modelled |
6.5 |
22 |
Fold:Barrel-sandwich hybrid
Superfamily:Ribosomal L27 protein-like
Family:ECR1 N-terminal domain-like |
| 63 | c3eiwA_ |
|
not modelled |
6.2 |
8 |
PDB header:transport protein
Chain: A: PDB Molecule:htsa protein;
PDBTitle: crystal structure of staphylococcus aureus lipoprotein, htsa
|
| 64 | c1mp9B_ |
|
not modelled |
6.1 |
21 |
PDB header:dna binding protein
Chain: B: PDB Molecule:tata-binding protein;
PDBTitle: tbp from a mesothermophilic archaeon, sulfolobus2 acidocaldarius
|
| 65 | c3frnA_ |
|
not modelled |
6.1 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:flagellar protein flga;
PDBTitle: crystal structure of flagellar protein flga from thermotoga maritima2 msb8
|
| 66 | d2q07a1 |
|
not modelled |
6.0 |
12 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:PUA domain |
| 67 | c2eh0A_ |
|
not modelled |
5.9 |
30 |
PDB header:transport protein
Chain: A: PDB Molecule:kinesin-like protein kif1b;
PDBTitle: solution structure of the fha domain from human kinesin-2 like protein kif1b
|
| 68 | c3nnkC_ |
|
not modelled |
5.7 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:ureidoglycine-glyoxylate aminotransferase;
PDBTitle: biochemical and structural characterization of a ureidoglycine2 aminotransferase in the klebsiella pneumoniae uric acid catabolic3 pathway
|
| 69 | d2q8oa1 |
|
not modelled |
5.7 |
38 |
Fold:TNF-like
Superfamily:TNF-like
Family:TNF-like |
| 70 | c1d3uA_ |
|
not modelled |
5.6 |
16 |
PDB header:gene regulation/dna
Chain: A: PDB Molecule:tata-binding protein;
PDBTitle: tata-binding protein/transcription factor (ii)b/bre+tata-2 box complex from pyrococcus woesei
|
| 71 | d1hmja_ |
|
not modelled |
5.6 |
38 |
Fold:RPB5-like RNA polymerase subunit
Superfamily:RPB5-like RNA polymerase subunit
Family:RPB5 |
| 72 | c1w9qB_ |
|
not modelled |
5.5 |
21 |
PDB header:cell adhesion
Chain: B: PDB Molecule:syntenin 1;
PDBTitle: crystal structure of the pdz tandem of human syntenin in2 complex with tnefaf peptide
|
| 73 | c2r3yC_ |
|
not modelled |
5.4 |
18 |
PDB header:hydrolase/hydrolase activator
Chain: C: PDB Molecule:protease degs;
PDBTitle: crystal structure of the degs protease in complex with the2 ywf activating peptide
|
| 74 | c3mb8A_ |
|
not modelled |
5.3 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of purine nucleoside phosphorylase from toxoplasma2 gondii in complex with immucillin-h
|
| 75 | d1qe5a_ |
|
not modelled |
5.3 |
12 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 76 | d1o6aa_ |
|
not modelled |
5.2 |
10 |
Fold:Surface presentation of antigens (SPOA)
Superfamily:Surface presentation of antigens (SPOA)
Family:Surface presentation of antigens (SPOA) |