| 1 |
|
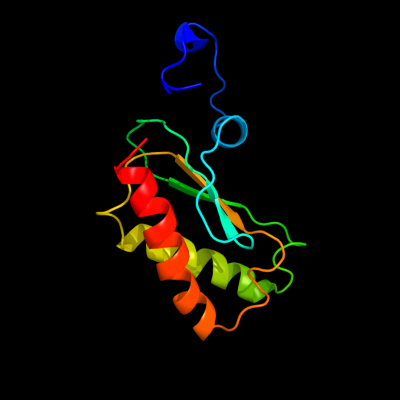
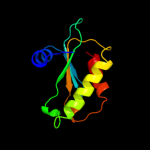
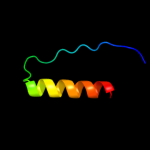
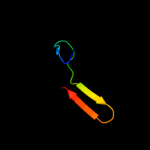
PDB 2ljp chain A
Region: 1 - 119
Aligned: 119
Modelled: 119
Confidence: 100.0%
Identity: 100%
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease p protein component;
PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for e.coli2 ribonuclease p protein
Phyre2
| 2 |
|
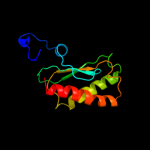
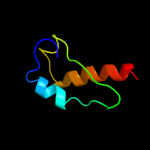
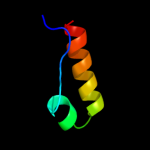
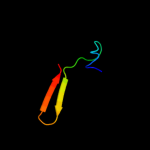
PDB 1d6t chain A
Region: 4 - 112
Aligned: 108
Modelled: 109
Confidence: 100.0%
Identity: 24%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: RNase P protein
Phyre2
| 3 |
|
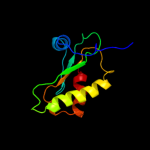
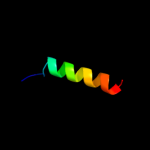
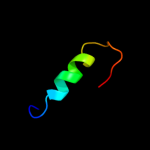
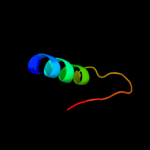
PDB 1a6f chain A
Region: 6 - 112
Aligned: 106
Modelled: 107
Confidence: 100.0%
Identity: 28%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: RNase P protein
Phyre2
| 4 |
|
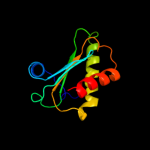
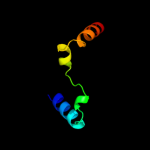
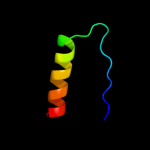
PDB 1nz0 chain A
Region: 10 - 112
Aligned: 102
Modelled: 103
Confidence: 100.0%
Identity: 29%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: RNase P protein
Phyre2
| 5 |
|
PDB 2c4r chain L
Region: 51 - 113
Aligned: 48
Modelled: 49
Confidence: 38.5%
Identity: 10%
PDB header:hydrolase
Chain: L: PDB Molecule:ribonuclease e;
PDBTitle: catalytic domain of e. coli rnase e
Phyre2
| 6 |
|
PDB 1q6o chain A
Region: 82 - 113
Aligned: 32
Modelled: 32
Confidence: 15.9%
Identity: 0%
Fold: TIM beta/alpha-barrel
Superfamily: Ribulose-phoshate binding barrel
Family: Decarboxylase
Phyre2
| 7 |
|
PDB 2y94 chain C
Region: 89 - 109
Aligned: 21
Modelled: 21
Confidence: 11.7%
Identity: 14%
PDB header:transferase
Chain: C: PDB Molecule:5'-amp-activated protein kinase catalytic subunit alpha-1;
PDBTitle: structure of an active form of mammalian ampk
Phyre2
| 8 |
|
PDB 3oqv chain A
Region: 63 - 110
Aligned: 47
Modelled: 48
Confidence: 11.0%
Identity: 13%
PDB header:protein binding
Chain: A: PDB Molecule:albc;
PDBTitle: albc, a cyclodipeptide synthase from streptomyces noursei
Phyre2
| 9 |
|
PDB 1qjh chain A
Region: 80 - 111
Aligned: 32
Modelled: 32
Confidence: 10.1%
Identity: 6%
Fold: Ferredoxin-like
Superfamily: Ribosomal protein S6
Family: Ribosomal protein S6
Phyre2
| 10 |
|
PDB 3bvj chain A
Region: 82 - 113
Aligned: 32
Modelled: 32
Confidence: 9.7%
Identity: 16%
PDB header:lyase
Chain: A: PDB Molecule:uridine 5'-monophosphate synthase;
PDBTitle: crystal structure of human orotidine 5'-monophosphate decarboxylase2 complexed with xmp
Phyre2
| 11 |
|
PDB 1tte chain A
Region: 63 - 85
Aligned: 23
Modelled: 23
Confidence: 9.4%
Identity: 17%
PDB header:ligase
Chain: A: PDB Molecule:ubiquitin-conjugating enzyme e2-24 kda;
PDBTitle: the structure of a class ii ubiquitin-conjugating enzyme,2 ubc1.
Phyre2
| 12 |
|
PDB 2kjw chain A
Region: 83 - 111
Aligned: 29
Modelled: 29
Confidence: 8.6%
Identity: 7%
PDB header:ribosomal protein
Chain: A: PDB Molecule:30s ribosomal protein s6;
PDBTitle: solution structure and backbone dynamics of the permutant2 p54-55
Phyre2
| 13 |
|
PDB 2qcn chain A
Region: 82 - 113
Aligned: 32
Modelled: 32
Confidence: 8.5%
Identity: 16%
PDB header:lyase
Chain: A: PDB Molecule:uridine 5'-monophosphate synthase;
PDBTitle: covalent complex of the orotidine-5'-monophosphate decarboxylase2 domain of human ump synthase with 6-iodo-ump
Phyre2
| 14 |
|
PDB 2j5a chain A domain 1
Region: 76 - 111
Aligned: 36
Modelled: 36
Confidence: 8.1%
Identity: 11%
Fold: Ferredoxin-like
Superfamily: Ribosomal protein S6
Family: Ribosomal protein S6
Phyre2
| 15 |
|
PDB 1lou chain A
Region: 80 - 111
Aligned: 32
Modelled: 32
Confidence: 7.7%
Identity: 6%
Fold: Ferredoxin-like
Superfamily: Ribosomal protein S6
Family: Ribosomal protein S6
Phyre2
| 16 |
|
PDB 3bbn chain F
Region: 80 - 109
Aligned: 30
Modelled: 30
Confidence: 6.6%
Identity: 17%
PDB header:ribosome
Chain: F: PDB Molecule:ribosomal protein s6;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
Phyre2
| 17 |
|
PDB 1vsr chain A
Region: 64 - 87
Aligned: 24
Modelled: 24
Confidence: 6.5%
Identity: 42%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: Very short patch repair (VSR) endonuclease
Phyre2
| 18 |
|
PDB 2fsu chain A domain 1
Region: 71 - 97
Aligned: 27
Modelled: 27
Confidence: 6.4%
Identity: 15%
Fold: PLP-dependent transferase-like
Superfamily: PhnH-like
Family: PhnH-like
Phyre2
| 19 |
|
PDB 2fsu chain A
Region: 71 - 97
Aligned: 27
Modelled: 27
Confidence: 6.4%
Identity: 15%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein phnh;
PDBTitle: crystal structure of the phnh protein from escherichia coli
Phyre2
| 20 |
|
PDB 2ayv chain A domain 1
Region: 64 - 87
Aligned: 24
Modelled: 24
Confidence: 5.4%
Identity: 17%
Fold: UBC-like
Superfamily: UBC-like
Family: UBC-related
Phyre2
| 21 |
|
| 22 |
|