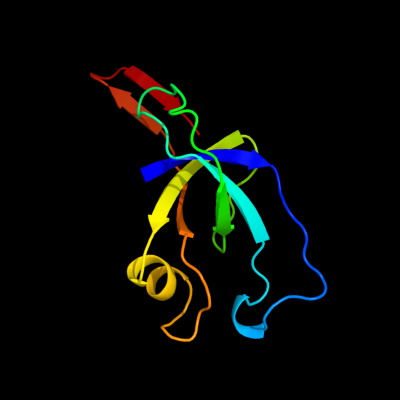
| 1 | d2qw7a1
|
|
 |
100.0 |
39 |
Fold:OB-fold
Superfamily:EutN/CcmL-like
Family:EutN/CcmL-like |
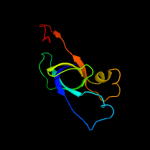
| 2 | d2hd3a1
|
|
 |
100.0 |
97 |
Fold:OB-fold
Superfamily:EutN/CcmL-like
Family:EutN/CcmL-like |
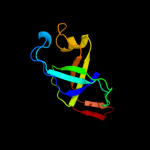
| 3 | d2rcfa1
|
|
 |
100.0 |
41 |
Fold:OB-fold
Superfamily:EutN/CcmL-like
Family:EutN/CcmL-like |
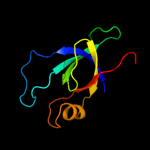
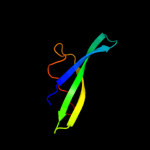
| 4 | d1jjcb3
|
|
 |
36.5 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Myf domain |
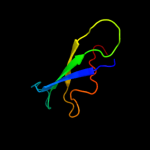
| 5 | c2akwB_
|
|
 |
29.7 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:phenylalanyl-trna synthetase beta chain;
PDBTitle: crystal structure of t.thermophilus phenylalanyl-trna synthetase2 complexed with p-cl-phenylalanine
|
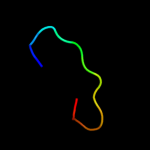
| 6 | d2z1ca1
|
|
 |
20.5 |
29 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
| 7 | c3bbnQ_
|
|
 |
19.4 |
16 |
PDB header:ribosome
Chain: Q: PDB Molecule:ribosomal protein s17;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
|
| 8 | c3bu2B_
|
|
 |
17.0 |
18 |
PDB header:rna binding protein
Chain: B: PDB Molecule:putative trna-binding protein;
PDBTitle: crystal structure of a trna-binding protein from2 staphylococcus saprophyticus subsp. saprophyticus.3 northeast structural genomics consortium target syr77
|
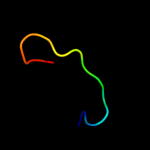
| 9 | d2ot2a1
|
|
 |
16.7 |
36 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
| 10 | d2gy9q1
|
|
 |
15.8 |
14 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 11 | d2z1ea2
|
|
 |
15.5 |
38 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
| 12 | c3d3rA_
|
|
 |
13.8 |
29 |
PDB header:chaperone
Chain: A: PDB Molecule:hydrogenase assembly chaperone hypc/hupf;
PDBTitle: crystal structure of the hydrogenase assembly chaperone hypc/hupf2 family protein from shewanella oneidensis mr-1
|
| 13 | c1rfoC_
|
|
 |
13.8 |
55 |
PDB header:viral protein
Chain: C: PDB Molecule:whisker antigen control protein;
PDBTitle: trimeric foldon of the t4 phagehead fibritin
|
| 14 | d2id1a1
|
|
 |
13.1 |
20 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Iojap/YbeB-like |
| 15 | d3d3ra1
|
|
 |
12.9 |
21 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
| 16 | d2zoda2
|
|
 |
11.2 |
31 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
| 17 | c3balB_
|
|
 |
10.9 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:acetylacetone-cleaving enzyme;
PDBTitle: crystal structure of an acetylacetone dioxygenase from2 acinetobacter johnsonii
|
| 18 | d1e8ca2
|
|
 |
10.7 |
19 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 19 | d2jfga2
|
|
 |
10.2 |
24 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 20 | d1g2914
|
|
 |
9.4 |
22 |
Fold:OB-fold
Superfamily:MOP-like
Family:ABC-transporter additional domain |
| 21 | c1avyA_ |
|
not modelled |
7.7 |
55 |
PDB header:coiled coil
Chain: A: PDB Molecule:fibritin;
PDBTitle: fibritin deletion mutant m (bacteriophage t4)
|
| 22 | c1ox3A_ |
|
not modelled |
7.5 |
55 |
PDB header:chaperone
Chain: A: PDB Molecule:fibritin;
PDBTitle: crystal structure of mini-fibritin
|
| 23 | d1wpga1 |
|
not modelled |
7.3 |
28 |
Fold:Double-stranded beta-helix
Superfamily:Calcium ATPase, transduction domain A
Family:Calcium ATPase, transduction domain A |
| 24 | d3c9ua2 |
|
not modelled |
7.2 |
38 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
| 25 | c3mvnA_ |
|
not modelled |
7.2 |
31 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate:l-alanyl-gamma-d-glutamayl-medo-
PDBTitle: crystal structure of a domain from a putative udp-n-acetylmuramate:l-2 alanyl-gamma-d-glutamayl-medo-diaminopimelate ligase from haemophilus3 ducreyi 35000hp
|
| 26 | d1c0aa2 |
|
not modelled |
7.1 |
15 |
Fold:DCoH-like
Superfamily:GAD domain-like
Family:GAD domain |
| 27 | c1e8cB_ |
|
not modelled |
7.0 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:udp-n-acetylmuramoylalanyl-d-glutamate--2,6-
PDBTitle: structure of mure the udp-n-acetylmuramyl tripeptide2 synthetase from e. coli
|
| 28 | c3b8eC_ |
|
not modelled |
6.9 |
31 |
PDB header:hydrolase/transport protein
Chain: C: PDB Molecule:sodium/potassium-transporting atpase subunit
PDBTitle: crystal structure of the sodium-potassium pump
|
| 29 | d1vk3a3 |
|
not modelled |
6.8 |
17 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
| 30 | c3upsA_ |
|
not modelled |
6.6 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:iojap-like protein;
PDBTitle: crystal structure of iojap-like protein from zymomonas mobilis
|
| 31 | d1ylqa1 |
|
not modelled |
6.5 |
20 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Catalytic subunit of bi-partite nucleotidyltransferase |
| 32 | d2atza1 |
|
not modelled |
6.3 |
25 |
Fold:Prim-pol domain
Superfamily:Prim-pol domain
Family:HP0184-like |
| 33 | c2opkC_ |
|
not modelled |
6.1 |
70 |
PDB header:isomerase
Chain: C: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a putative mannose-6-phosphate isomerase2 (reut_a1446) from ralstonia eutropha jmp134 at 2.10 a resolution
|
| 34 | d2c42a3 |
|
not modelled |
6.0 |
35 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Pyruvate-ferredoxin oxidoreductase, PFOR, domain II |
| 35 | c1s1hQ_ |
|
not modelled |
5.9 |
13 |
PDB header:ribosome
Chain: Q: PDB Molecule:40s ribosomal protein s11;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
|
| 36 | c3b9bA_ |
|
not modelled |
5.9 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: structure of the e2 beryllium fluoride complex of the serca2 ca2+-atpase
|
| 37 | c3fq6A_ |
|
not modelled |
5.6 |
40 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: the crystal structure of a methyltransferase domain from bacteroides2 thetaiotaomicron vpi
|
| 38 | c3ixzA_ |
|
not modelled |
5.6 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:potassium-transporting atpase alpha;
PDBTitle: pig gastric h+/k+-atpase complexed with aluminium fluoride
|
| 39 | c3p9zA_ |
|
not modelled |
5.2 |
12 |
PDB header:ligase
Chain: A: PDB Molecule:uroporphyrinogen iii cosynthase (hemd);
PDBTitle: crystal structure of uroporphyrinogen-iii synthetase from helicobacter2 pylori 26695
|
| 40 | c3ajvA_ |
|
not modelled |
5.2 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: splicing endonuclease from aeropyrum pernix
|
| 41 | c1v1hB_ |
|
not modelled |
5.1 |
50 |
PDB header:adenovirus
Chain: B: PDB Molecule:fibritin, fiber protein;
PDBTitle: adenovirus fibre shaft sequence n-terminally fused to the2 bacteriophage t4 fibritin foldon trimerisation motif with3 a short linker
|
| 42 | c2gq1A_ |
|
not modelled |
5.1 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:fructose-1,6-bisphosphatase;
PDBTitle: crystal structure of recombinant type i fructose-1,6-bisphosphatase2 from escherichia coli complexed with sulfate ions
|
| 43 | d2o5aa1 |
|
not modelled |
5.1 |
24 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Iojap/YbeB-like |