| 1 | c2jlhA_ |
|
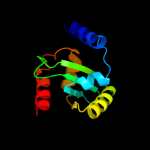 |
100.0 |
34 |
PDB header:protein transport
Chain: A: PDB Molecule:yop proteins translocation protein u;
PDBTitle: crystal structure of the cytoplasmic domain of yersinia2 pestis yscu n263a mutant
|
| 2 | c3bzrA_ |
|
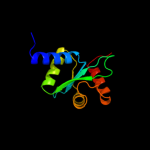 |
100.0 |
28 |
PDB header:membrane protein, protein transport
Chain: A: PDB Molecule:escu;
PDBTitle: crystal structure of escu c-terminal domain with n262d mutation, space2 group p 41 21 2
|
| 3 | d3bzra1 |
|
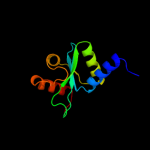 |
100.0 |
28 |
Fold:EscU C-terminal domain-like
Superfamily:EscU C-terminal domain-like
Family:EscU C-terminal domain-like |
| 4 | c3t7yB_ |
|
 |
100.0 |
31 |
PDB header:protein transport
Chain: B: PDB Molecule:yop proteins translocation protein u;
PDBTitle: structure of an autocleavage-inactive mutant of the cytoplasmic domain2 of ct091, the yscu homologue of chlamydia trachomatis
|
| 5 | c3c01H_ |
|
 |
99.9 |
20 |
PDB header:membrane protein, protein transport
Chain: H: PDB Molecule:surface presentation of antigens protein spas;
PDBTitle: crystal structural of native spas c-terminal domain
|
| 6 | c3c00B_ |
|
 |
99.9 |
30 |
PDB header:membrane protein, protein transport
Chain: B: PDB Molecule:escu;
PDBTitle: crystal structural of the mutated g247t escu/spas c-terminal domain
|
| 7 | c2vt1B_ |
|
 |
99.9 |
23 |
PDB header:membrane protein
Chain: B: PDB Molecule:surface presentation of antigens protein spas;
PDBTitle: crystal structure of the cytoplasmic domain of spa40, the2 specificity switch for the shigella flexneri type iii3 secretion system
|
| 8 | c2yvqA_ |
|
 |
87.1 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoyl-phosphate synthase;
PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
|
| 9 | d1a0rp_ |
|
 |
76.9 |
17 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Phosducin |
| 10 | d1wo8a1 |
|
 |
75.8 |
20 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
| 11 | d2nu7b1 |
|
 |
73.4 |
17 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 12 | d1a9xa2 |
|
 |
64.9 |
13 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain |
| 13 | d1uk5a_ |
|
 |
62.6 |
24 |
Fold:Spectrin repeat-like
Superfamily:BAG domain
Family:BAG domain |
| 14 | d1ugoa_ |
|
 |
62.5 |
14 |
Fold:Spectrin repeat-like
Superfamily:BAG domain
Family:BAG domain |
| 15 | c3peuB_ |
|
 |
62.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:nucleoporin gle1;
PDBTitle: s. cerevisiae dbp5 l327v c-terminal domain bound to gle1 h337r and ip6
|
| 16 | d1m7ka_ |
|
 |
57.9 |
10 |
Fold:Spectrin repeat-like
Superfamily:BAG domain
Family:BAG domain |
| 17 | c1yj7A_ |
|
 |
57.0 |
17 |
PDB header:protein transport
Chain: A: PDB Molecule:escj;
PDBTitle: crystal structure of enteropathogenic e.coli (epec) type iii secretion2 system protein escj
|
| 18 | d1d8ja_ |
|
 |
56.0 |
10 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:The central core domain of TFIIE beta |
| 19 | c3a8yD_ |
|
 |
53.9 |
24 |
PDB header:hydrolase
Chain: D: PDB Molecule:bag family molecular chaperone regulator 5;
PDBTitle: crystal structure of the complex between the bag5 bd5 and2 hsp70 nbd
|
| 20 | d1rv3a_ |
|
 |
53.6 |
34 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 21 | d1eucb1 |
|
not modelled |
50.1 |
17 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 22 | d1mv8a2 |
|
not modelled |
49.8 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 23 | c1r6xA_ |
|
not modelled |
49.5 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:atp:sulfate adenylyltransferase;
PDBTitle: the crystal structure of a truncated form of yeast atp2 sulfurylase, lacking the c-terminal aps kinase-like domain,3 in complex with sulfate
|
| 24 | d2a7va1 |
|
not modelled |
49.1 |
34 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 25 | c2a7vA_ |
|
not modelled |
49.1 |
34 |
PDB header:transferase
Chain: A: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: human mitochondrial serine hydroxymethyltransferase 2
|
| 26 | d1ko7a1 |
|
not modelled |
48.5 |
12 |
Fold:MurF and HprK N-domain-like
Superfamily:HprK N-terminal domain-like
Family:HPr kinase/phoshatase HprK N-terminal domain |
| 27 | d1g8fa2 |
|
not modelled |
47.8 |
24 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:ATP sulfurylase catalytic domain |
| 28 | c2o14A_ |
|
not modelled |
47.2 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein yxim;
PDBTitle: x-ray crystal structure of protein yxim_bacsu from bacillus2 subtilis. northeast structural genomics consortium target3 sr595
|
| 29 | c2vycA_ |
|
not modelled |
46.8 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:biodegradative arginine decarboxylase;
PDBTitle: crystal structure of acid induced arginine decarboxylase2 from e. coli
|
| 30 | d2csua3 |
|
not modelled |
44.8 |
27 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 31 | d1t9ba1 |
|
not modelled |
43.5 |
13 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 32 | d1knxa1 |
|
not modelled |
43.4 |
15 |
Fold:MurF and HprK N-domain-like
Superfamily:HprK N-terminal domain-like
Family:HPr kinase/phoshatase HprK N-terminal domain |
| 33 | c3nysA_ |
|
not modelled |
41.2 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase wbpe;
PDBTitle: x-ray structure of the k185a mutant of wbpe (wlbe) from pseudomonas2 aeruginosa in complex with plp at 1.45 angstrom resolution
|
| 34 | d1bj4a_ |
|
not modelled |
37.6 |
29 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 35 | c1c4kA_ |
|
not modelled |
37.5 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:protein (ornithine decarboxylase);
PDBTitle: ornithine decarboxylase mutant (gly121tyr)
|
| 36 | c2r18A_ |
|
not modelled |
37.2 |
26 |
PDB header:viral protein
Chain: A: PDB Molecule:capsid assembly protein vp3;
PDBTitle: structural insights into the multifunctional protein vp3 of2 birnaviruses
|
| 37 | d1oi7a1 |
|
not modelled |
36.7 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 38 | c3iynO_ |
|
not modelled |
35.9 |
24 |
PDB header:virus
Chain: O: PDB Molecule:hexon-associated protein;
PDBTitle: 3.6-angstrom cryoem structure of human adenovirus type 5
|
| 39 | c2qjfB_ |
|
not modelled |
35.0 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional 3'-phosphoadenosine 5'-
PDBTitle: crystal structure of atp-sulfurylase domain of human paps2 synthetase 1
|
| 40 | c3bcxA_ |
|
not modelled |
34.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:cdp-6-deoxy-l-threo-d-glycero-4-hexulose-3-
PDBTitle: e1 dehydrase
|
| 41 | d2nu7a1 |
|
not modelled |
33.8 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 42 | d1gqoa_ |
|
not modelled |
33.1 |
34 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 43 | d1f6ga_ |
|
not modelled |
32.9 |
13 |
Fold:Voltage-gated potassium channels
Superfamily:Voltage-gated potassium channels
Family:Voltage-gated potassium channels |
| 44 | d1vmda_ |
|
not modelled |
32.7 |
14 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
| 45 | c3me5A_ |
|
not modelled |
31.9 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:cytosine-specific methyltransferase;
PDBTitle: crystal structure of putative dna cytosine methylase from shigella2 flexneri 2a str. 301
|
| 46 | d1qzxa2 |
|
not modelled |
31.3 |
35 |
Fold:Signal peptide-binding domain
Superfamily:Signal peptide-binding domain
Family:Signal peptide-binding domain |
| 47 | c1yrwA_ |
|
not modelled |
30.6 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:protein arna;
PDBTitle: crystal structure of e.coli arna transformylase domain
|
| 48 | c3e6gA_ |
|
not modelled |
30.2 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:cystathionine gamma-lyase-like protein;
PDBTitle: crystal structure of xometc, a cystathionine c-lyase-like2 protein from xanthomonas oryzae pv.oryzae
|
| 49 | d2naca2 |
|
not modelled |
29.5 |
27 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 50 | c3dm5A_ |
|
not modelled |
29.5 |
19 |
PDB header:rna binding protein, transport protein
Chain: A: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: structures of srp54 and srp19, the two proteins assembling2 the ribonucleic core of the signal recognition particle3 from the archaeon pyrococcus furiosus.
|
| 51 | d1c4ka2 |
|
not modelled |
28.7 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Ornithine decarboxylase major domain |
| 52 | d1ydwa1 |
|
not modelled |
28.6 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 53 | d1vl2a1 |
|
not modelled |
28.5 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 54 | c2po3B_ |
|
not modelled |
28.4 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:4-dehydrase;
PDBTitle: crystal structure analysis of desi in the presence of its2 tdp-sugar product
|
| 55 | c3n8kG_ |
|
not modelled |
28.1 |
39 |
PDB header:lyase
Chain: G: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: type ii dehydroquinase from mycobacterium tuberculosis complexed with2 citrazinic acid
|
| 56 | d1ni5a1 |
|
not modelled |
28.0 |
6 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PP-loop ATPase |
| 57 | c2jqeA_ |
|
not modelled |
27.6 |
25 |
PDB header:signaling protein
Chain: A: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: soution structure of af54 m-domain
|
| 58 | c3qi6B_ |
|
not modelled |
27.5 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:cystathionine gamma-synthase metb (cgs);
PDBTitle: crystal structure of cystathionine gamma-synthase metb (cgs) from2 mycobacterium ulcerans agy99
|
| 59 | d1cs1a_ |
|
not modelled |
27.5 |
12 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 60 | d1wy5a1 |
|
not modelled |
27.4 |
16 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:PP-loop ATPase |
| 61 | c3h7uA_ |
|
not modelled |
27.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldo-keto reductase;
PDBTitle: crystal structure of the plant stress-response enzyme akr4c9
|
| 62 | d1d2ta_ |
|
not modelled |
27.0 |
8 |
Fold:Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily:Acid phosphatase/Vanadium-dependent haloperoxidase
Family:Type 2 phosphatidic acid phosphatase, PAP2 |
| 63 | c2uygF_ |
|
not modelled |
26.6 |
43 |
PDB header:lyase
Chain: F: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystallogaphic structure of the typeii 3-dehydroquinase2 from thermus thermophilus
|
| 64 | d1j20a1 |
|
not modelled |
26.4 |
8 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 65 | c2csuB_ |
|
not modelled |
26.4 |
27 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:457aa long hypothetical protein;
PDBTitle: crystal structure of ph0766 from pyrococcus horikoshii ot3
|
| 66 | c3f4lF_ |
|
not modelled |
26.4 |
12 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:putative oxidoreductase yhhx;
PDBTitle: crystal structure of a probable oxidoreductase yhhx in2 triclinic form. northeast structural genomics target er647
|
| 67 | d1hq1a_ |
|
not modelled |
26.4 |
20 |
Fold:Signal peptide-binding domain
Superfamily:Signal peptide-binding domain
Family:Signal peptide-binding domain |
| 68 | c3nt5B_ |
|
not modelled |
26.3 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inositol 2-dehydrogenase/d-chiro-inositol 3-dehydrogenase;
PDBTitle: crystal structure of myo-inositol dehydrogenase from bacillus subtilis2 with bound cofactor and product inosose
|
| 69 | c2c7tA_ |
|
not modelled |
26.3 |
35 |
PDB header:transferase
Chain: A: PDB Molecule:glutamine-2-deoxy-scyllo-inosose
PDBTitle: crystal structure of the plp-bound form of btrr,2 a dual functional aminotransferase involved in butirosin3 biosynthesis.
|
| 70 | c3dc1A_ |
|
not modelled |
26.2 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:kynurenine/alpha-aminoadipate aminotransferase
PDBTitle: crystal structure of kynurenine aminotransferase ii complex with2 alpha-ketoglutarate
|
| 71 | c1wziA_ |
|
not modelled |
26.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: structural basis for alteration of cofactor specificity of2 malate dehydrogenase from thermus flavus
|
| 72 | d2ffha2 |
|
not modelled |
25.9 |
25 |
Fold:Signal peptide-binding domain
Superfamily:Signal peptide-binding domain
Family:Signal peptide-binding domain |
| 73 | d1ekqa_ |
|
not modelled |
25.9 |
19 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Thiamin biosynthesis kinases |
| 74 | c3u80A_ |
|
not modelled |
25.7 |
53 |
PDB header:unknown function
Chain: A: PDB Molecule:3-dehydroquinate dehydratase, type ii;
PDBTitle: 1.60 angstrom resolution crystal structure of a 3-dehydroquinate2 dehydratase-like protein from bifidobacterium longum
|
| 75 | d1uqra_ |
|
not modelled |
25.7 |
43 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 76 | c2hroA_ |
|
not modelled |
25.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of the full-lenght enzyme i of the pts system from2 staphylococcus carnosus
|
| 77 | c1a5zA_ |
|
not modelled |
25.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: lactate dehydrogenase from thermotoga maritima (tmldh)
|
| 78 | c3aemD_ |
|
not modelled |
25.0 |
9 |
PDB header:lyase
Chain: D: PDB Molecule:methionine gamma-lyase;
PDBTitle: reaction intermediate structure of entamoeba histolytica methionine2 gamma-lyase 1 containing michaelis complex and methionine imine-3 pyridoxamine-5'-phosphate
|
| 79 | c3dpiA_ |
|
not modelled |
24.5 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:nad+ synthetase;
PDBTitle: crystal structure of nad+ synthetase from burkholderia pseudomallei
|
| 80 | d1h05a_ |
|
not modelled |
24.5 |
39 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 81 | c3ndnC_ |
|
not modelled |
24.3 |
6 |
PDB header:lyase
Chain: C: PDB Molecule:o-succinylhomoserine sulfhydrylase;
PDBTitle: crystal structure of o-succinylhomoserine sulfhydrylase from2 mycobacterium tuberculosis covalently bound to pyridoxal-5-phosphate
|
| 82 | d2ji7a1 |
|
not modelled |
23.8 |
23 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 83 | c2lbwA_ |
|
not modelled |
23.2 |
22 |
PDB header:rna binding protein
Chain: A: PDB Molecule:h/aca ribonucleoprotein complex subunit 2;
PDBTitle: solution structure of the s. cerevisiae h/aca rnp protein nhp2p-s82w2 mutant
|
| 84 | c2y8nC_ |
|
not modelled |
22.8 |
19 |
PDB header:lyase
Chain: C: PDB Molecule:4-hydroxyphenylacetate decarboxylase large subunit;
PDBTitle: crystal structure of glycyl radical enzyme
|
| 85 | c2cb1A_ |
|
not modelled |
22.7 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:o-acetyl homoserine sulfhydrylase;
PDBTitle: crystal structure of o-actetyl homoserine sulfhydrylase2 from thermus thermophilus hb8,oah2.
|
| 86 | c1zh8B_ |
|
not modelled |
22.7 |
8 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of oxidoreductase (tm0312) from thermotoga maritima2 at 2.50 a resolution
|
| 87 | c2bn5A_ |
|
not modelled |
22.7 |
11 |
PDB header:nuclear protein
Chain: A: PDB Molecule:psi;
PDBTitle: p-element somatic inhibitor protein complex with u1-70k2 proline-rich peptide
|
| 88 | d1ui5a2 |
|
not modelled |
22.6 |
21 |
Fold:Tetracyclin repressor-like, C-terminal domain
Superfamily:Tetracyclin repressor-like, C-terminal domain
Family:Tetracyclin repressor-like, C-terminal domain |
| 89 | c3lwzC_ |
|
not modelled |
22.4 |
34 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.65 angstrom resolution crystal structure of type ii 3-2 dehydroquinate dehydratase (aroq) from yersinia pestis
|
| 90 | d1gtza_ |
|
not modelled |
22.4 |
47 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 91 | c2r0tA_ |
|
not modelled |
22.3 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:pyridoxamine 5-phosphate-dependent dehydrase;
PDBTitle: crystal sructure of gdp-4-keto-6-deoxymannose-3-dehydratase2 with a trapped plp-glutamate geminal diamine
|
| 92 | d2c4va1 |
|
not modelled |
22.1 |
25 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 93 | c3c1aB_ |
|
not modelled |
22.1 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase (zp_00056571.1) from2 magnetospirillum magnetotacticum ms-1 at 1.85 a resolution
|
| 94 | c3qh9A_ |
|
not modelled |
21.7 |
13 |
PDB header:structural protein
Chain: A: PDB Molecule:liprin-beta-2;
PDBTitle: human liprin-beta2 coiled-coil
|
| 95 | c3c19A_ |
|
not modelled |
21.7 |
38 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein mk0293;
PDBTitle: crystal structure of protein mk0293 from methanopyrus kandleri av19
|
| 96 | d1rlga_ |
|
not modelled |
21.5 |
20 |
Fold:Bacillus chorismate mutase-like
Superfamily:L30e-like
Family:L30e/L7ae ribosomal proteins |
| 97 | c2f3oB_ |
|
not modelled |
21.5 |
17 |
PDB header:unknown function
Chain: B: PDB Molecule:pyruvate formate-lyase 2;
PDBTitle: crystal structure of a glycyl radical enzyme from archaeoglobus2 fulgidus
|
| 98 | d1ibja_ |
|
not modelled |
21.5 |
18 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 99 | c1ibjC_ |
|
not modelled |
21.5 |
18 |
PDB header:lyase
Chain: C: PDB Molecule:cystathionine beta-lyase;
PDBTitle: crystal structure of cystathionine beta-lyase from arabidopsis2 thaliana
|
| 100 | d1b9ha_ |
|
not modelled |
21.4 |
27 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 101 | d1p3da1 |
|
not modelled |
21.3 |
7 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 102 | c3iayA_ |
|
not modelled |
21.1 |
20 |
PDB header:transferase/dna
Chain: A: PDB Molecule:dna polymerase delta catalytic subunit;
PDBTitle: ternary complex of dna polymerase delta
|
| 103 | d2a6ta2 |
|
not modelled |
21.1 |
19 |
Fold:Nudix
Superfamily:Nudix
Family:mRNA decapping enzyme-like |
| 104 | c2pq1B_ |
|
not modelled |
20.9 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:ap4a hydrolase;
PDBTitle: crystal structure of ap4a hydrolase complexed with amp and2 atp (aq_158) from aquifex aeolicus vf5
|
| 105 | c3ezyB_ |
|
not modelled |
20.7 |
17 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure of probable dehydrogenase tm_0414 from2 thermotoga maritima
|
| 106 | c2k9iB_ |
|
not modelled |
20.6 |
38 |
PDB header:dna binding protein
Chain: B: PDB Molecule:plasmid prn1, complete sequence;
PDBTitle: nmr structure of plasmid copy control protein orf56 from2 sulfolobus islandicus
|
| 107 | c2ogeC_ |
|
not modelled |
20.3 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:transaminase;
PDBTitle: x-ray structure of s. venezuelae desv in its internal2 aldimine form
|

