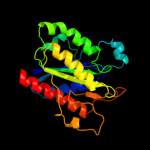
| 1 | c3b6iB_ |
|
 |
100.0 |
100 |
PDB header:flavoprotein
Chain: B: PDB Molecule:flavoprotein wrba;
PDBTitle: wrba from escherichia coli, native structure
|
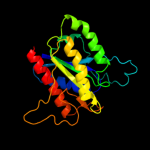
| 2 | c2zkiH_ |
|
 |
100.0 |
40 |
PDB header:transcription
Chain: H: PDB Molecule:199aa long hypothetical trp repressor binding
PDBTitle: crystal structure of hypothetical trp repressor binding2 protein from sul folobus tokodaii (st0872)
|
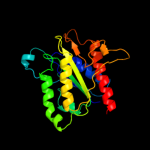
| 3 | d1ydga_ |
|
 |
100.0 |
38 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
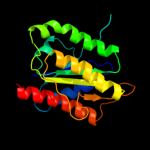
| 4 | d2a5la1 |
|
 |
100.0 |
43 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 5 | c3d7nA_ |
|
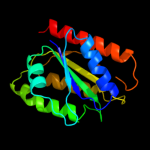 |
100.0 |
31 |
PDB header:electron transport
Chain: A: PDB Molecule:flavodoxin, wrba-like protein;
PDBTitle: the crystal structure of the flavodoxin, wrba-like protein from2 agrobacterium tumefaciens
|
| 6 | d2arka1 |
|
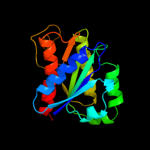 |
100.0 |
32 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 7 | d1sqsa_ |
|
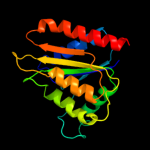 |
100.0 |
13 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Hypothetical protein SP1951 |
| 8 | c2q62A_ |
|
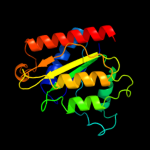 |
100.0 |
17 |
PDB header:flavoprotein
Chain: A: PDB Molecule:arsh;
PDBTitle: crystal structure of arsh from sinorhizobium meliloti
|
| 9 | d1nni1_ |
|
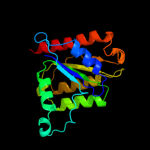 |
99.9 |
15 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:NADPH-dependent FMN reductase |
| 10 | c3lcmB_ |
|
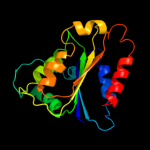 |
99.9 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of smu.1420 from streptococcus mutans ua159
|
| 11 | d1ycga1 |
|
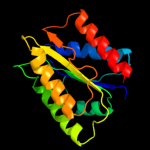 |
99.9 |
22 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 12 | d1rtta_ |
|
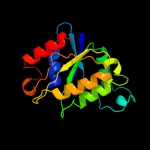 |
99.9 |
13 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:NADPH-dependent FMN reductase |
| 13 | d1t5ba_ |
|
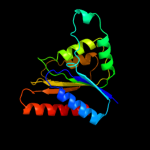 |
99.9 |
22 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 14 | c2fzvC_ |
|
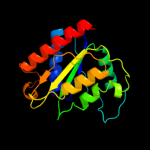 |
99.9 |
13 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:putative arsenical resistance protein;
PDBTitle: crystal structure of an apo form of a flavin-binding protein from2 shigella flexneri
|
| 15 | c3k1yE_ |
|
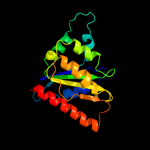 |
99.9 |
18 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:oxidoreductase;
PDBTitle: x-ray structure of oxidoreductase from corynebacterium2 diphtheriae. orthorombic crystal form, northeast structural3 genomics consortium target cdr100d
|
| 16 | d1t0ia_ |
|
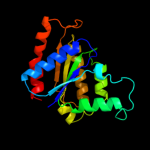 |
99.9 |
17 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:NADPH-dependent FMN reductase |
| 17 | d1e5da1 |
|
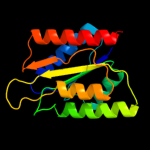 |
99.9 |
25 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 18 | c3fniA_ |
|
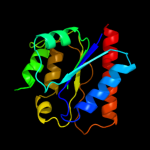 |
99.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative diflavin flavoprotein a 3;
PDBTitle: crystal structure of a diflavin flavoprotein a3 (all3895) from nostoc2 sp., northeast structural genomics consortium target nsr431a
|
| 19 | c2hpvA_ |
|
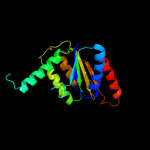 |
99.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fmn-dependent nadh-azoreductase;
PDBTitle: crystal structure of fmn-dependent azoreductase from enterococcus2 faecalis
|
| 20 | c2vzhA_ |
|
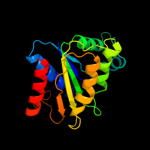 |
99.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh-dependent fmn reductase;
PDBTitle: structures of nadh:fmn oxidoreductase (emob)-fmn complex
|
| 21 | c1ychD_ |
|
not modelled |
99.9 |
22 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nitric oxide reductase;
PDBTitle: x-ray crystal structures of moorella thermoacetica fpra.2 novel diiron site structure and mechanistic insights into3 a scavenging nitric oxide reductase
|
| 22 | c3fvwA_ |
|
not modelled |
99.9 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative nad(p)h-dependent fmn reductase;
PDBTitle: crystal structure of the q8dwd8_strmu protein from2 streptococcus mutans. northeast structural genomics3 consortium target smr99.
|
| 23 | c2ohiB_ |
|
not modelled |
99.9 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:type a flavoprotein fpra;
PDBTitle: crystal structure of coenzyme f420h2 oxidase (fpra), a diiron2 flavoprotein, reduced state
|
| 24 | c2q9uB_ |
|
not modelled |
99.9 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:a-type flavoprotein;
PDBTitle: crystal structure of the flavodiiron protein from giardia2 intestinalis
|
| 25 | d2fzva1 |
|
not modelled |
99.9 |
13 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:NADPH-dependent FMN reductase |
| 26 | d2qwxa1 |
|
not modelled |
99.9 |
18 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 27 | c3p0rA_ |
|
not modelled |
99.9 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:azoreductase;
PDBTitle: crystal structure of azoreductase from bacillus anthracis str. sterne
|
| 28 | d2z98a1 |
|
not modelled |
99.9 |
24 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 29 | d1qrda_ |
|
not modelled |
99.9 |
16 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 30 | c3hlyA_ |
|
not modelled |
99.9 |
22 |
PDB header:flavoprotein
Chain: A: PDB Molecule:flavodoxin-like domain;
PDBTitle: crystal structure of the flavodoxin-like domain from2 synechococcus sp q5mzp6_synp6 protein. northeast structural3 genomics consortium target snr135d.
|
| 31 | d1d4aa_ |
|
not modelled |
99.9 |
17 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 32 | d1vmea1 |
|
not modelled |
99.9 |
22 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 33 | c2v9cA_ |
|
not modelled |
99.9 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fmn-dependent nadh-azoreductase 1;
PDBTitle: x-ray crystallographic structure of a pseudomonas2 aeruginosa azoreductase in complex with methyl red.
|
| 34 | d1dxqa_ |
|
not modelled |
99.9 |
22 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 35 | d1rlia_ |
|
not modelled |
99.9 |
21 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Hypothetical protein YwqN |
| 36 | c1e5dA_ |
|
not modelled |
99.9 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:rubredoxin\:oxygen oxidoreductase;
PDBTitle: rubredoxin oxygen:oxidoreductase (roo) from anaerobe2 desulfovibrio gigas
|
| 37 | c1vmeB_ |
|
not modelled |
99.9 |
21 |
PDB header:electron transport
Chain: B: PDB Molecule:flavoprotein;
PDBTitle: crystal structure of flavoprotein (tm0755) from thermotoga maritima at2 1.80 a resolution
|
| 38 | c3f2vA_ |
|
not modelled |
99.9 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:general stress protein 14;
PDBTitle: crystal structure of the general stress protein 142 (tde0354) in complex with fmn from treponema denticola,3 northeast structural genomics consortium target tdr58.
|
| 39 | c3edoA_ |
|
not modelled |
99.8 |
21 |
PDB header:flavoprotein
Chain: A: PDB Molecule:putative trp repressor binding protein;
PDBTitle: crystal structure of flavoprotein in complex with fmn2 (yp_193882.1) from lactobacillus acidophilus ncfm at 1.203 a resolution
|
| 40 | c3klbA_ |
|
not modelled |
99.8 |
21 |
PDB header:flavoprotein
Chain: A: PDB Molecule:putative flavoprotein;
PDBTitle: crystal structure of putative flavoprotein in complex with fmn2 (yp_213683.1) from bacteroides fragilis nctc 9343 at 1.75 a3 resolution
|
| 41 | c3rpeA_ |
|
not modelled |
99.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:modulator of drug activity b;
PDBTitle: 1.1 angstrom crystal structure of putative modulator of drug activity2 (mdab) from yersinia pestis co92.
|
| 42 | d2fz5a1 |
|
not modelled |
99.8 |
24 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 43 | d5nula_ |
|
not modelled |
99.8 |
24 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 44 | c2amjD_ |
|
not modelled |
99.7 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:modulator of drug activity b;
PDBTitle: crystal structure of modulator of drug activity b from escherichia2 coli o157:h7
|
| 45 | c3f6sI_ |
|
not modelled |
99.7 |
24 |
PDB header:electron transport
Chain: I: PDB Molecule:flavodoxin;
PDBTitle: desulfovibrio desulfuricans (atcc 29577) oxidized flavodoxin2 alternate conformers
|
| 46 | c3ha2B_ |
|
not modelled |
99.7 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadph-quinone reductase;
PDBTitle: crystal structure of protein (nadph-quinone reductase) from2 p.pentosaceus, northeast structural genomics consortium target ptr24a
|
| 47 | c3eywA_ |
|
not modelled |
99.7 |
19 |
PDB header:transport protein
Chain: A: PDB Molecule:c-terminal domain of glutathione-regulated potassium-efflux
PDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
|
| 48 | c2wc1A_ |
|
not modelled |
99.6 |
18 |
PDB header:electron transport
Chain: A: PDB Molecule:flavodoxin;
PDBTitle: three-dimensional structure of the nitrogen fixation2 flavodoxin (niff) from rhodobacter capsulatus at 2.2 a
|
| 49 | d1ag9a_ |
|
not modelled |
99.6 |
19 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 50 | d1oboa_ |
|
not modelled |
99.6 |
18 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 51 | c2hnbA_ |
|
not modelled |
99.6 |
15 |
PDB header:electron transport
Chain: A: PDB Molecule:protein mioc;
PDBTitle: solution structure of a bacterial holo-flavodoxin
|
| 52 | d1czna_ |
|
not modelled |
99.6 |
18 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 53 | d1f4pa_ |
|
not modelled |
99.5 |
23 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 54 | d1yoba1 |
|
not modelled |
99.5 |
18 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 55 | d2fcra_ |
|
not modelled |
99.5 |
20 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 56 | d1ykga1 |
|
not modelled |
99.5 |
19 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Cytochrome p450 reductase N-terminal domain-like |
| 57 | d1fuea_ |
|
not modelled |
99.4 |
16 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 58 | c1bvyF_ |
|
not modelled |
99.4 |
22 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:protein (cytochrome p450 bm-3);
PDBTitle: complex of the heme and fmn-binding domains of the2 cytochrome p450(bm-3)
|
| 59 | d1bvyf_ |
|
not modelled |
99.4 |
22 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 60 | d1b1ca_ |
|
not modelled |
99.4 |
19 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Cytochrome p450 reductase N-terminal domain-like |
| 61 | d1tlla2 |
|
not modelled |
99.4 |
18 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Cytochrome p450 reductase N-terminal domain-like |
| 62 | c3hr4C_ |
|
not modelled |
99.3 |
16 |
PDB header:oxidoreductase/metal binding protein
Chain: C: PDB Molecule:nitric oxide synthase, inducible;
PDBTitle: human inos reductase and calmodulin complex
|
| 63 | d1ja1a2 |
|
not modelled |
99.2 |
19 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Cytochrome p450 reductase N-terminal domain-like |
| 64 | c1tllA_ |
|
not modelled |
98.9 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nitric-oxide synthase, brain;
PDBTitle: crystal structure of rat neuronal nitric-oxide synthase2 reductase module at 2.3 a resolution.
|
| 65 | c2bpoA_ |
|
not modelled |
98.8 |
21 |
PDB header:reductase
Chain: A: PDB Molecule:nadph-cytochrom p450 reductase;
PDBTitle: crystal structure of the yeast cpr triple mutant: d74g,2 y75f, k78a.
|
| 66 | c2x2oA_ |
|
not modelled |
98.7 |
22 |
PDB header:flavoprotein
Chain: A: PDB Molecule:nrdi protein;
PDBTitle: the flavoprotein nrdi from bacillus cereus with the2 initially oxidized fmn cofactor in an intermediate3 radiation reduced state
|
| 67 | c1j9zB_ |
|
not modelled |
98.6 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadph-cytochrome p450 reductase;
PDBTitle: cypor-w677g
|
| 68 | d1rlja_ |
|
not modelled |
98.3 |
19 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavoprotein NrdI |
| 69 | c3n39D_ |
|
not modelled |
98.1 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:protein nrdi;
PDBTitle: ribonucleotide reductase dimanganese(ii)-nrdf from escherichia coli in2 complex with nrdi
|
| 70 | c2kyrA_ |
|
not modelled |
95.1 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:fructose-like phosphotransferase enzyme iib component 1;
PDBTitle: solution structure of enzyme iib subunit of pts system from2 escherichia coli k12. northeast structural genomics consortium target3 er315/ontario center for structural proteomics target ec0544
|
| 71 | c3czcA_ |
|
not modelled |
95.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:rmpb;
PDBTitle: the crystal structure of a putative pts iib(ptxb) from2 streptococcus mutans
|
| 72 | d2r4qa1 |
|
not modelled |
94.7 |
33 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Fructose specific IIB subunit-like |
| 73 | c3a9rA_ |
|
not modelled |
94.3 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:d-arabinose isomerase;
PDBTitle: x-ray structures of bacillus pallidus d-arabinose2 isomerasecomplex with (4r)-2-methylpentane-2,4-diol
|
| 74 | d1n1ea2 |
|
not modelled |
92.1 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 75 | d2a9va1 |
|
not modelled |
91.9 |
12 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 76 | d1uqra_ |
|
not modelled |
91.8 |
25 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 77 | c3kklA_ |
|
not modelled |
91.7 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable chaperone protein hsp33;
PDBTitle: crystal structure of functionally unknown hsp33 from2 saccharomyces cerevisiae
|
| 78 | c1tvmA_ |
|
not modelled |
91.3 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, galactitol-specific iib component;
PDBTitle: nmr structure of enzyme gatb of the galactitol-specific2 phosphoenolpyruvate-dependent phosphotransferase system
|
| 79 | d2r48a1 |
|
not modelled |
90.7 |
27 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Fructose specific IIB subunit-like |
| 80 | c3ghyA_ |
|
not modelled |
90.7 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ketopantoate reductase protein;
PDBTitle: crystal structure of a putative ketopantoate reductase from ralstonia2 solanacearum molk2
|
| 81 | c3c7cB_ |
|
not modelled |
90.5 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:octopine dehydrogenase;
PDBTitle: a structural basis for substrate and stereo selectivity in2 octopine dehydrogenase (odh-nadh-l-arginine)
|
| 82 | c3u80A_ |
|
not modelled |
90.5 |
20 |
PDB header:unknown function
Chain: A: PDB Molecule:3-dehydroquinate dehydratase, type ii;
PDBTitle: 1.60 angstrom resolution crystal structure of a 3-dehydroquinate2 dehydratase-like protein from bifidobacterium longum
|
| 83 | d1jvna2 |
|
not modelled |
90.5 |
22 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 84 | c3d54D_ |
|
not modelled |
90.2 |
21 |
PDB header:ligase
Chain: D: PDB Molecule:phosphoribosylformylglycinamidine synthase 1;
PDBTitle: stucture of purlqs from thermotoga maritima
|
| 85 | c2gcgB_ |
|
not modelled |
90.2 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyoxylate reductase/hydroxypyruvate reductase;
PDBTitle: ternary crystal structure of human glyoxylate2 reductase/hydroxypyruvate reductase
|
| 86 | c2vpiA_ |
|
not modelled |
90.1 |
24 |
PDB header:ligase
Chain: A: PDB Molecule:gmp synthase;
PDBTitle: human gmp synthetase - glutaminase domain
|
| 87 | c1jvnB_ |
|
not modelled |
90.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional histidine biosynthesis protein hishf;
PDBTitle: crystal structure of imidazole glycerol phosphate synthase: a tunnel2 through a (beta/alpha)8 barrel joins two active sites
|
| 88 | c3qvjB_ |
|
not modelled |
89.9 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:putative hydantoin racemase;
PDBTitle: allantoin racemase from klebsiella pneumoniae
|
| 89 | c3dojA_ |
|
not modelled |
89.6 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dehydrogenase-like protein;
PDBTitle: structure of glyoxylate reductase 1 from arabidopsis2 (atglyr1)
|
| 90 | c2ho3D_ |
|
not modelled |
89.5 |
11 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of oxidoreductase, gfo/idh/moca family from2 streptococcus pneumoniae
|
| 91 | d1txga2 |
|
not modelled |
89.3 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 92 | c3ic5A_ |
|
not modelled |
88.8 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative saccharopine dehydrogenase;
PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
|
| 93 | c3dhnA_ |
|
not modelled |
88.7 |
16 |
PDB header:isomerase, lyase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of the putative epimerase q89z24_bactn2 from bacteroides thetaiotaomicron. northeast structural3 genomics consortium target btr310.
|
| 94 | c3nbmA_ |
|
not modelled |
87.7 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, lactose-specific iibc components;
PDBTitle: the lactose-specific iib component domain structure of the2 phosphoenolpyruvate:carbohydrate phosphotransferase system (pts) from3 streptococcus pneumoniae.
|
| 95 | c2l2qA_ |
|
not modelled |
87.2 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, cellobiose-specific iib component (cela);
PDBTitle: solution structure of cellobiose-specific phosphotransferase iib2 component protein from borrelia burgdorferi
|
| 96 | c3s40C_ |
|
not modelled |
86.6 |
23 |
PDB header:transferase
Chain: C: PDB Molecule:diacylglycerol kinase;
PDBTitle: the crystal structure of a diacylglycerol kinases from bacillus2 anthracis str. sterne
|
| 97 | c3euwB_ |
|
not modelled |
86.3 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:myo-inositol dehydrogenase;
PDBTitle: crystal structure of a myo-inositol dehydrogenase from corynebacterium2 glutamicum atcc 13032
|
| 98 | d2p1ra1 |
|
not modelled |
85.5 |
15 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
| 99 | c2axqA_ |
|
not modelled |
84.8 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase;
PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
|
| 100 | d2ab0a1 |
|
not modelled |
84.7 |
17 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:DJ-1/PfpI |
| 101 | c3upsA_ |
|
not modelled |
84.5 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:iojap-like protein;
PDBTitle: crystal structure of iojap-like protein from zymomonas mobilis
|
| 102 | c3f5dA_ |
|
not modelled |
84.3 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein ydea;
PDBTitle: crystal structure of a protein of unknown function from2 bacillus subtilis
|
| 103 | d1u7za_ |
|
not modelled |
84.2 |
14 |
Fold:Ribokinase-like
Superfamily:CoaB-like
Family:CoaB-like |
| 104 | c3plnA_ |
|
not modelled |
83.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-glucose 6-dehydrogenase;
PDBTitle: crystal structure of klebsiella pneumoniae udp-glucose 6-dehydrogenase2 complexed with udp-glucose
|
| 105 | d2nv0a1 |
|
not modelled |
83.9 |
32 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 106 | c3d8tB_ |
|
not modelled |
83.9 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: thermus thermophilus uroporphyrinogen iii synthase
|
| 107 | d1iiba_ |
|
not modelled |
83.8 |
15 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Lactose/Cellobiose specific IIB subunit |
| 108 | d1ka9h_ |
|
not modelled |
83.7 |
26 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 109 | d1ks9a2 |
|
not modelled |
83.7 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 110 | c3l6dB_ |
|
not modelled |
83.3 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase from pseudomonas putida2 kt2440
|
| 111 | d1u9ca_ |
|
not modelled |
82.6 |
16 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:DJ-1/PfpI |
| 112 | d1mv8a3 |
|
not modelled |
82.6 |
20 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:UDP-glucose/GDP-mannose dehydrogenase C-terminal domain
Family:UDP-glucose/GDP-mannose dehydrogenase C-terminal domain |
| 113 | c2f1kD_ |
|
not modelled |
82.5 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of synechocystis arogenate dehydrogenase
|
| 114 | c3ktdC_ |
|
not modelled |
81.6 |
8 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of a putative prephenate dehydrogenase (cgl0226)2 from corynebacterium glutamicum atcc 13032 at 2.60 a resolution
|
| 115 | c3fd8A_ |
|
not modelled |
81.5 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
| 116 | d1vjta1 |
|
not modelled |
81.4 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 117 | c2z2vA_ |
|
not modelled |
81.2 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical protein ph1688;
PDBTitle: crystal structure of l-lysine dehydrogenase from2 hyperthermophilic archaeon pyrococcus horikoshii
|
| 118 | d1i36a2 |
|
not modelled |
81.1 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 119 | c3ia7A_ |
|
not modelled |
80.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:calg4;
PDBTitle: crystal structure of calg4, the calicheamicin glycosyltransferase
|
| 120 | c2eklA_ |
|
not modelled |
80.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: structure of st1218 protein from sulfolobus tokodaii
|

