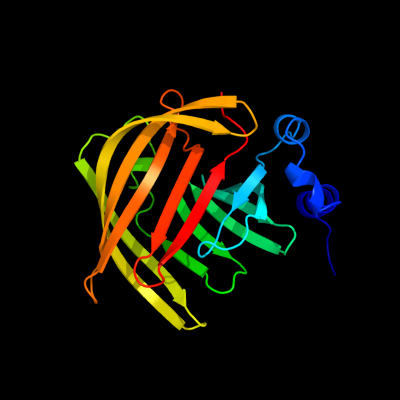
1 d2in5a1
100.0
96
Fold: YmcC-likeSuperfamily: YmcC-likeFamily: YmcC-like2 d1bmlc3
44.1
27
Fold: beta-Grasp (ubiquitin-like)Superfamily: Staphylokinase/streptokinaseFamily: Staphylokinase/streptokinase3 d2byoa1
41.3
16
Fold: LolA-like prokaryotic lipoproteins and lipoprotein localization factorsSuperfamily: Prokaryotic lipoproteins and lipoprotein localization factorsFamily: LppX-like4 c3jslA_
22.1
18
PDB header: ligaseChain: A: PDB Molecule: dna ligase;PDBTitle: crystal structure of the adenylation domain of nad+-2 dependent dna ligase from staphylococcus aureus
5 c2x6vB_
18.5
28
PDB header: transcription/dnaChain: B: PDB Molecule: t-box transcription factor tbx5;PDBTitle: crystal structure of human tbx5 in the dna-bound and dna-2 free form
6 d1a0ia1
18.4
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: DNA ligase/mRNA capping enzyme postcatalytic domain7 c3o0lB_
15.6
25
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a pfam duf1425 family member (shew_1734) from2 shewanella sp. pv-4 at 1.81 a resolution
8 c3fzxA_
12.6
12
PDB header: lipid binding proteinChain: A: PDB Molecule: putative exported protein;PDBTitle: crystal structure of a putative exported protein with ymcc-like fold2 (bf2203) from bacteroides fragilis nctc 9343 at 2.22 a resolution
9 d1v9pa3
12.5
24
Fold: ATP-graspSuperfamily: DNA ligase/mRNA capping enzyme, catalytic domainFamily: Adenylation domain of NAD+-dependent DNA ligase10 d1ogmx1
12.4
16
Fold: Dextranase, N-terminal domainSuperfamily: Dextranase, N-terminal domainFamily: Dextranase, N-terminal domain11 c3kdvB_
12.2
12
PDB header: dna binding proteinChain: B: PDB Molecule: dna damage response b protein;PDBTitle: crystal structure of dna damage response b (ddrb) from deinococcus2 geothermalis
12 c2v43A_
11.8
15
PDB header: regulatorChain: A: PDB Molecule: sigma-e factor regulatory protein rseb;PDBTitle: crystal structure of rseb: a sensor for periplasmic stress2 response in e. coli
13 c2h8bA_
11.3
57
PDB header: hormone/growth factorChain: A: PDB Molecule: insulin-like 3;PDBTitle: solution structure of insl3
14 c2k6tA_
11.3
57
PDB header: hormoneChain: A: PDB Molecule: insulin-like 3 a chain;PDBTitle: solution structure of the relaxin-like factor
15 c2k6uA_
10.9
57
PDB header: hormoneChain: A: PDB Molecule: insulin-like 3 a chain;PDBTitle: the solution structure of a conformationally restricted2 fully active derivative of the human relaxin-like factor3 (rlf)
16 d1h6fa_
9.5
24
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: T-box17 c3mlqE_
9.1
19
PDB header: transferase/transcriptionChain: E: PDB Molecule: transcription-repair coupling factor;PDBTitle: crystal structure of the thermus thermophilus transcription-repair2 coupling factor rna polymerase interacting domain with the thermus3 aquaticus rna polymerase beta1 domain
18 d1zbsa2
9.0
27
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like19 c3mlqH_
7.7
19
PDB header: transferase/transcriptionChain: H: PDB Molecule: transcription-repair coupling factor;PDBTitle: crystal structure of the thermus thermophilus transcription-repair2 coupling factor rna polymerase interacting domain with the thermus3 aquaticus rna polymerase beta1 domain
20 d1dgsa3
7.2
18
Fold: ATP-graspSuperfamily: DNA ligase/mRNA capping enzyme, catalytic domainFamily: Adenylation domain of NAD+-dependent DNA ligase21 c3bacA_
not modelled
6.8
29
PDB header: ligaseChain: A: PDB Molecule: dna ligase;PDBTitle: structural basis for the inhibition of bacterial nad+2 dependent dna ligase
22 d1bhia_
not modelled
6.8
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H223 d2f1la2
not modelled
6.8
8
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: RimM N-terminal domain-like24 d1b04a_
not modelled
6.5
18
Fold: ATP-graspSuperfamily: DNA ligase/mRNA capping enzyme, catalytic domainFamily: Adenylation domain of NAD+-dependent DNA ligase25 d1azwa_
not modelled
5.9
21
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Proline iminopeptidase-like26 d4tsva_
not modelled
5.9
17
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like27 d1r57a_
not modelled
5.8
12
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT28 c3o44G_
not modelled
5.8
16
PDB header: toxinChain: G: PDB Molecule: hemolysin;PDBTitle: crystal structure of the vibrio cholerae cytolysin (hlya) heptameric2 pore
29 c1c9lA_
not modelled
5.8
44
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: clathrin;PDBTitle: peptide-in-groove interactions link target proteins to the2 b-propeller of clathrin
30 d2cupa3
not modelled
5.8
44
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain31 d1a9xb1
not modelled
5.7
20
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Carbamoyl phosphate synthetase, small subunit N-terminal domainFamily: Carbamoyl phosphate synthetase, small subunit N-terminal domain32 c2j8pA_
not modelled
5.6
36
PDB header: nuclear proteinChain: A: PDB Molecule: cleavage stimulation factor 64 kda subunit;PDBTitle: nmr structure of c-terminal domain of human cstf-64
33 d1utca2
not modelled
5.5
44
Fold: 7-bladed beta-propellerSuperfamily: Clathrin heavy-chain terminal domainFamily: Clathrin heavy-chain terminal domain34 c3f6zB_
not modelled
5.4
14
PDB header: hydrolaseChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of pseudomonas aeruginosa mlic in complex2 with hen egg white lysozyme
35 d3bl9a2
not modelled
5.4
24
Fold: mRNA decapping enzyme DcpS N-terminal domainSuperfamily: mRNA decapping enzyme DcpS N-terminal domainFamily: mRNA decapping enzyme DcpS N-terminal domain36 d1vlra2
not modelled
5.3
18
Fold: mRNA decapping enzyme DcpS N-terminal domainSuperfamily: mRNA decapping enzyme DcpS N-terminal domainFamily: mRNA decapping enzyme DcpS N-terminal domain