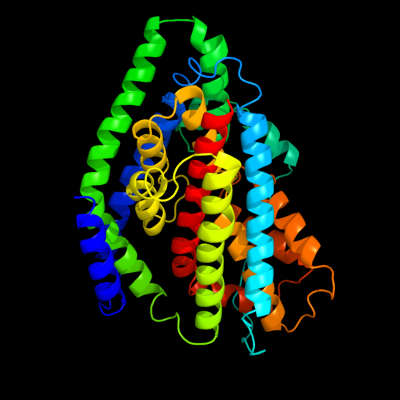
| 1 | d2nwwa1
|
|
 |
100.0 |
26 |
Fold:Proton glutamate symport protein
Superfamily:Proton glutamate symport protein
Family:Proton glutamate symport protein |
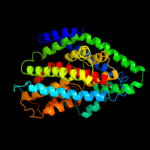
| 2 | d1iwga8
|
|
 |
58.6 |
14 |
Fold:Multidrug efflux transporter AcrB transmembrane domain
Superfamily:Multidrug efflux transporter AcrB transmembrane domain
Family:Multidrug efflux transporter AcrB transmembrane domain |
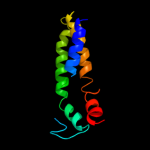
| 3 | d1a77a1
|
|
 |
22.0 |
11 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
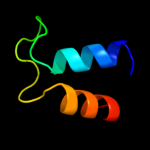
| 4 | c1oy8A_
|
|
 |
18.9 |
13 |
PDB header:membrane protein
Chain: A: PDB Molecule:acriflavine resistance protein b;
PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
|
| 5 | c3ah5E_
|
|
 |
18.7 |
19 |
PDB header:transferase
Chain: E: PDB Molecule:thymidylate synthase thyx;
PDBTitle: crystal structure of flavin dependent thymidylate synthase thyx from2 helicobacter pylori complexed with fad and dump
|
| 6 | d1o26a_
|
|
 |
16.0 |
14 |
Fold:Thymidylate synthase-complementing protein Thy1
Superfamily:Thymidylate synthase-complementing protein Thy1
Family:Thymidylate synthase-complementing protein Thy1 |
| 7 | d1bboa1
|
|
 |
12.3 |
9 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 8 | c3k07A_
|
|
 |
10.4 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:cation efflux system protein cusa;
PDBTitle: crystal structure of cusa
|
| 9 | d1mc8a1
|
|
 |
9.2 |
11 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
| 10 | c2cmyB_
|
|
 |
8.9 |
50 |
PDB header:hydrolase
Chain: B: PDB Molecule:veronica hederifolia trypsin inhibitor;
PDBTitle: crystal complex between bovine trypsin and veronica2 hederifolia trypsin inhibitor
|
| 11 | d2cmyb1
|
|
 |
8.9 |
50 |
Fold:Toxic hairpin
Superfamily:VhTI-like
Family:VhTI-like |
| 12 | d1rxwa1
|
|
 |
8.6 |
11 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
| 13 | d1kyza1
|
|
 |
8.4 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Plant O-methyltransferase, N-terminal domain |
| 14 | c2cfaB_
|
|
 |
8.3 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:thymidylate synthase;
PDBTitle: structure of viral flavin-dependant thymidylate synthase2 thyx
|
| 15 | d1ul1x1
|
|
 |
8.2 |
5 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
| 16 | c2e75E_
|
|
 |
8.1 |
28 |
PDB header:photosynthesis
Chain: E: PDB Molecule:cytochrome b6-f complex subunit 6;
PDBTitle: crystal structure of the cytochrome b6f complex with 2-nonyl-4-2 hydroxyquinoline n-oxide (nqno) from m.laminosus
|
| 17 | c1vf5R_
|
|
 |
8.1 |
28 |
PDB header:photosynthesis
Chain: R: PDB Molecule:protein pet l;
PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
|
| 18 | c2e74E_
|
|
 |
8.1 |
28 |
PDB header:photosynthesis
Chain: E: PDB Molecule:cytochrome b6-f complex subunit 6;
PDBTitle: crystal structure of the cytochrome b6f complex from m.laminosus
|
| 19 | c1vf5E_
|
|
 |
8.1 |
28 |
PDB header:photosynthesis
Chain: E: PDB Molecule:protein pet l;
PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
|
| 20 | d2e74e1
|
|
 |
8.1 |
28 |
Fold:Single transmembrane helix
Superfamily:PetL subunit of the cytochrome b6f complex
Family:PetL subunit of the cytochrome b6f complex |
| 21 | c2e76E_ |
|
not modelled |
8.1 |
28 |
PDB header:photosynthesis
Chain: E: PDB Molecule:cytochrome b6-f complex subunit 6;
PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
|
| 22 | d1xo1a1 |
|
not modelled |
8.0 |
18 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
| 23 | d1fp1d1 |
|
not modelled |
7.2 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Plant O-methyltransferase, N-terminal domain |
| 24 | d2eiaa2 |
|
not modelled |
7.1 |
10 |
Fold:Retrovirus capsid protein, N-terminal core domain
Superfamily:Retrovirus capsid protein, N-terminal core domain
Family:Retrovirus capsid protein, N-terminal core domain |
| 25 | d1b43a1 |
|
not modelled |
6.8 |
11 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
| 26 | c1tujA_ |
|
not modelled |
6.5 |
22 |
PDB header:transport protein
Chain: A: PDB Molecule:odorant binding protein asp2;
PDBTitle: solution structure of the honey bee general odorant binding2 protein asp2 in complex with trimethylsilyl-d4 propionate
|
| 27 | c3dl8D_ |
|
not modelled |
6.4 |
13 |
PDB header:protein transport
Chain: D: PDB Molecule:sece;
PDBTitle: structure of the complex of aquifex aeolicus secyeg and2 bacillus subtilis seca
|
| 28 | c2crnA_ |
|
not modelled |
6.3 |
10 |
PDB header:immune system
Chain: A: PDB Molecule:ubash3a protein;
PDBTitle: solution structure of the uba domain of human ubash3a2 protein
|
| 29 | d1iwga7 |
|
not modelled |
6.1 |
11 |
Fold:Multidrug efflux transporter AcrB transmembrane domain
Superfamily:Multidrug efflux transporter AcrB transmembrane domain
Family:Multidrug efflux transporter AcrB transmembrane domain |
| 30 | c2wlvA_ |
|
not modelled |
5.5 |
15 |
PDB header:virus protein
Chain: A: PDB Molecule:gag polyprotein;
PDBTitle: structure of the n-terminal capsid domain of hiv-2
|
| 31 | d1m9dc_ |
|
not modelled |
5.4 |
15 |
Fold:Retrovirus capsid protein, N-terminal core domain
Superfamily:Retrovirus capsid protein, N-terminal core domain
Family:Retrovirus capsid protein, N-terminal core domain |
| 32 | c2pcoA_ |
|
not modelled |
5.4 |
38 |
PDB header:toxin
Chain: A: PDB Molecule:latarcin-1;
PDBTitle: spatial structure and membrane permeabilization for2 latarcin-1, a spider antimicrobial peptide
|
| 33 | d2pxrc1 |
|
not modelled |
5.4 |
15 |
Fold:Retrovirus capsid protein, N-terminal core domain
Superfamily:Retrovirus capsid protein, N-terminal core domain
Family:Retrovirus capsid protein, N-terminal core domain |
| 34 | c2jwaA_ |
|
not modelled |
5.1 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
|