| 1 |
|
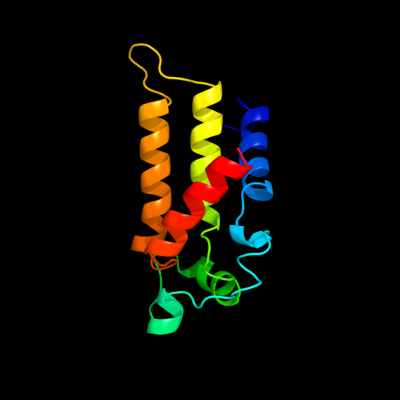
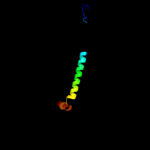
PDB 2akc chain C
Region: 69 - 223
Aligned: 119
Modelled: 124
Confidence: 99.9%
Identity: 23%
PDB header:hydrolase
Chain: C: PDB Molecule:class a nonspecific acid phosphatase phon;
PDBTitle: crystal structure of tungstate complex of the phon protein2 from s. typhimurium
Phyre2
| 2 |
|
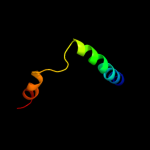
PDB 1d2t chain A
Region: 69 - 223
Aligned: 117
Modelled: 122
Confidence: 99.9%
Identity: 28%
Fold: Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily: Acid phosphatase/Vanadium-dependent haloperoxidase
Family: Type 2 phosphatidic acid phosphatase, PAP2
Phyre2
| 3 |
|
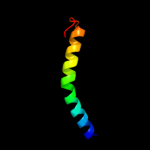
PDB 1vng chain A
Region: 77 - 211
Aligned: 105
Modelled: 110
Confidence: 98.3%
Identity: 21%
PDB header:haloperoxidase
Chain: A: PDB Molecule:vanadium chloroperoxidase;
PDBTitle: chloroperoxidase from the fungus curvularia inaequalis:2 mutant h404a
Phyre2
| 4 |
|
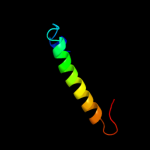
PDB 1vns chain A
Region: 77 - 211
Aligned: 105
Modelled: 110
Confidence: 98.3%
Identity: 22%
Fold: Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily: Acid phosphatase/Vanadium-dependent haloperoxidase
Family: Chloroperoxidase
Phyre2
| 5 |
|
PDB 1qi9 chain A
Region: 158 - 224
Aligned: 67
Modelled: 67
Confidence: 98.2%
Identity: 24%
Fold: Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily: Acid phosphatase/Vanadium-dependent haloperoxidase
Family: Haloperoxidase (bromoperoxidase)
Phyre2
| 6 |
|
PDB 1qhb chain A
Region: 130 - 224
Aligned: 95
Modelled: 95
Confidence: 97.6%
Identity: 16%
Fold: Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily: Acid phosphatase/Vanadium-dependent haloperoxidase
Family: Haloperoxidase (bromoperoxidase)
Phyre2
| 7 |
|
PDB 1up8 chain A
Region: 130 - 224
Aligned: 95
Modelled: 95
Confidence: 97.5%
Identity: 13%
Fold: Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily: Acid phosphatase/Vanadium-dependent haloperoxidase
Family: Haloperoxidase (bromoperoxidase)
Phyre2
| 8 |
|
PDB 1v54 chain D
Region: 201 - 252
Aligned: 52
Modelled: 52
Confidence: 63.8%
Identity: 15%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit IV
Family: Mitochondrial cytochrome c oxidase subunit IV
Phyre2
| 9 |
|
PDB 2y69 chain Q
Region: 201 - 252
Aligned: 52
Modelled: 52
Confidence: 63.8%
Identity: 15%
PDB header:electron transport
Chain: Q: PDB Molecule:cytochrome c oxidase subunit 4 isoform 1;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 10 |
|
PDB 2jln chain A
Region: 215 - 254
Aligned: 40
Modelled: 40
Confidence: 27.8%
Identity: 10%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 11 |
|
PDB 2a65 chain A domain 1
Region: 184 - 248
Aligned: 57
Modelled: 65
Confidence: 26.5%
Identity: 21%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 12 |
|
PDB 2knc chain B
Region: 212 - 253
Aligned: 42
Modelled: 42
Confidence: 23.7%
Identity: 12%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 13 |
|
PDB 2knc chain A
Region: 212 - 249
Aligned: 38
Modelled: 38
Confidence: 19.2%
Identity: 21%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 14 |
|
PDB 3qnq chain D
Region: 213 - 249
Aligned: 37
Modelled: 37
Confidence: 8.5%
Identity: 22%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 15 |
|
PDB 1l7v chain A
Region: 199 - 240
Aligned: 42
Modelled: 42
Confidence: 6.9%
Identity: 17%
Fold: ABC transporter involved in vitamin B12 uptake, BtuC
Superfamily: ABC transporter involved in vitamin B12 uptake, BtuC
Family: ABC transporter involved in vitamin B12 uptake, BtuC
Phyre2