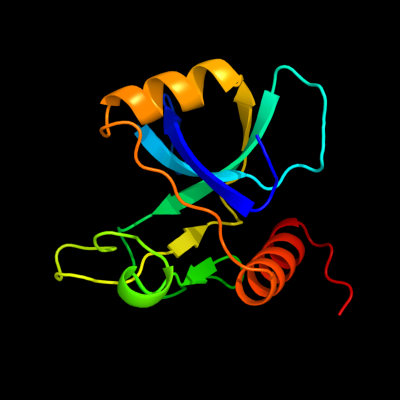
1 d1ne8a_
99.6
16
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK2 d1m1fa_
99.5
18
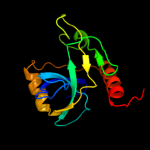
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK3 d1ub4a_
99.3
27
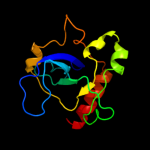
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK4 c1ym7C_
56.4
21
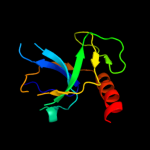
PDB header: transferaseChain: C: PDB Molecule: beta-adrenergic receptor kinase 1;PDBTitle: g protein-coupled receptor kinase 2 (grk2)
5 c3e7eA_
54.3
19
PDB header: transferaseChain: A: PDB Molecule: mitotic checkpoint serine/threonine-protein kinase bub1;PDBTitle: structure and substrate recruitment of the human spindle checkpoint2 kinase bub
6 c3qa8A_
38.9
18
PDB header: immune system, signaling proteinChain: A: PDB Molecule: mgc80376 protein;PDBTitle: crystal structure of inhibitor of kappa b kinase beta
7 c2kytA_
37.7
25
PDB header: hydrolaseChain: A: PDB Molecule: group xvi phospholipase a2;PDBTitle: solution struture of the h-rev107 n-terminal domain
8 c3pfqA_
35.5
21
PDB header: transferaseChain: A: PDB Molecule: protein kinase c beta type;PDBTitle: crystal structure and allosteric activation of protein kinase c beta2 ii
9 c3c4yA_
33.6
26
PDB header: transferaseChain: A: PDB Molecule: rhodopsin kinase;PDBTitle: crystal structure of apo form of g protein coupled receptor kinase 12 at 7.51a
10 d1kjwa2
25.3
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases11 c2a1aB_
24.2
16
PDB header: protein synthesis/transferaseChain: B: PDB Molecule: interferon-induced, double-stranded rna-activated proteinPDBTitle: pkr kinase domain-eif2alpha complex
12 c3iz5N_
15.9
30
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein l14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
13 d1fn9a_
15.9
23
Fold: Outer capsid protein sigma 3Superfamily: Outer capsid protein sigma 3Family: Outer capsid protein sigma 314 c3izcN_
14.8
25
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein rpl14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
15 c2jqmA_
12.9
33
PDB header: transferaseChain: A: PDB Molecule: envelope protein e;PDBTitle: yellow fever envelope protein domain iii nmr structure2 (s288-k398)
16 c4a19F_
12.7
25
PDB header: ribosomeChain: F: PDB Molecule: rpl14;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 2.
17 c2qg5D_
12.7
13
PDB header: transferaseChain: D: PDB Molecule: calcium/calmodulin-dependent protein kinase;PDBTitle: cryptosporidium parvum calcium dependent protein kinase cgd7_1840
18 d1qb5d_
12.5
24
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits19 d1x6oa1
11.8
25
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: eIF5a N-terminal domain-like20 c3dzoA_
11.2
22
PDB header: transferaseChain: A: PDB Molecule: rhoptry kinase domain;PDBTitle: crystal structure of a rhoptry kinase from toxoplasma gondii
21 c3byvA_
not modelled
10.5
23
PDB header: transferaseChain: A: PDB Molecule: rhoptry kinase;PDBTitle: crystal structure of toxoplasma gondii specific rhoptry2 antigen kinase domain
22 c1kjwA_
not modelled
10.4
14
PDB header: neuropeptideChain: A: PDB Molecule: postsynaptic density protein 95;PDBTitle: sh3-guanylate kinase module from psd-95
23 c3q60A_
not modelled
9.9
10
PDB header: transferaseChain: A: PDB Molecule: rop5b;PDBTitle: crystal structure of virulent allele rop5b pseudokinase domain bound2 to atp
24 d2je6i1
not modelled
9.6
27
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like25 d2joya1
not modelled
9.6
30
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal protein L14e26 c2acxB_
not modelled
8.9
21
PDB header: transferaseChain: B: PDB Molecule: g protein-coupled receptor kinase 6;PDBTitle: crystal structure of g protein coupled receptor kinase 6 bound to2 amppnp
27 d1qcfa3
not modelled
8.7
16
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: Protein kinases, catalytic subunit28 c3nahC_
not modelled
8.6
5
PDB header: transferaseChain: C: PDB Molecule: rna dependent rna polymerase;PDBTitle: crystal structures and functional analysis of murine norovirus rna-2 dependent rna polymerase
29 c4a18N_
not modelled
8.6
21
PDB header: ribosomeChain: N: PDB Molecule: rpl27;PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of molecule 1
30 c3ayhB_
not modelled
8.4
36
PDB header: transcriptionChain: B: PDB Molecule: dna-directed rna polymerase iii subunit rpc8;PDBTitle: crystal structure of the c17/25 subcomplex from s. pombe rna2 polymerase iii
31 c2vwiC_
not modelled
8.3
16
PDB header: transferaseChain: C: PDB Molecule: serine/threonine-protein kinase osr1;PDBTitle: structure of the osr1 kinase, a hypertension drug target
32 d2g4ca1
not modelled
8.1
12
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS33 c2b43D_
not modelled
8.1
4
PDB header: viral proteinChain: D: PDB Molecule: non-structural polyprotein;PDBTitle: crystal structure of the norwalk virus rna dependent rna polymerase2 from strain hu/nlv/dresden174/1997/ge
34 d1k8ib2
not modelled
7.7
50
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain35 c2cgvA_
not modelled
7.5
17
PDB header: transferaseChain: A: PDB Molecule: serine/threonine-protein kinase chk1;PDBTitle: identification of chemically diverse chk1 inhibitors by2 receptor-based virtual screening
36 c1khiA_
not modelled
7.4
16
PDB header: structural proteinChain: A: PDB Molecule: hex1;PDBTitle: crystal structure of hex1
37 d1qpca_
not modelled
7.4
13
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: Protein kinases, catalytic subunit38 d1u09a_
not modelled
7.3
9
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: RNA-dependent RNA-polymerase39 c2pziA_
not modelled
7.3
11
PDB header: transferaseChain: A: PDB Molecule: probable serine/threonine-protein kinase pkng;PDBTitle: crystal structure of protein kinase pkng from mycobacterium2 tuberculosis in complex with tetrahydrobenzothiophene ax20017
40 c3egpA_
not modelled
7.1
15
PDB header: viral proteinChain: A: PDB Molecule: envelope protein;PDBTitle: crystal structure analysis of dengue-1 envelope protein2 domain iii
41 c1imhD_
not modelled
7.1
16
PDB header: transcription/dnaChain: D: PDB Molecule: nuclear factor of activated t cells 5;PDBTitle: tonebp/dna complex
42 c2ckzB_
not modelled
7.1
43
PDB header: transferaseChain: B: PDB Molecule: dna-directed rna polymerase iii 25 kdPDBTitle: x-ray structure of rna polymerase iii subcomplex c17-c25.
43 c3ckxA_
not modelled
7.0
24
PDB header: transferaseChain: A: PDB Molecule: serine/threonine-protein kinase 24;PDBTitle: crystal structure of sterile 20-like kinase 3 (mst3, stk24)2 in complex with staurosporine
44 c1zp9A_
not modelled
6.8
5
PDB header: transferaseChain: A: PDB Molecule: rio1 kinase;PDBTitle: crystal structure of full-legnth a.fulgidus rio1 serine kinase bound2 to atp and mn2+ ions.
45 c2uutA_
not modelled
6.6
18
PDB header: hydrolaseChain: A: PDB Molecule: rna-directed rna polymerase;PDBTitle: the 2.4 angstrom resolution structure of the d346g mutant2 of the sapporo virus rdrp polymerase
46 c3uatA_
not modelled
6.5
15
PDB header: peptide binding proteinChain: A: PDB Molecule: disks large homolog 1;PDBTitle: guanylate kinase domains of the maguk family scaffold proteins as2 specific phospho-protein binding modules
47 c3lijA_
not modelled
6.5
22
PDB header: transferaseChain: A: PDB Molecule: calcium/calmodulin dependent protein kinase withPDBTitle: crystal structure of full length cpcdpk3 (cgd5_820) in2 complex with ca2+ and amppnp
48 c2ux8G_
not modelled
6.4
13
PDB header: transferaseChain: G: PDB Molecule: glucose-1-phosphate uridylyltransferase;PDBTitle: crystal structure of sphingomonas elodea atcc 31461 glucose-2 1-phosphate uridylyltransferase in complex with glucose-3 1-phosphate.
49 d1nsca_
not modelled
6.3
31
Fold: 6-bladed beta-propellerSuperfamily: SialidasesFamily: Sialidases (neuraminidases)50 c3f6kA_
not modelled
6.2
50
PDB header: signaling proteinChain: A: PDB Molecule: sortilin;PDBTitle: crystal structure of the vps10p domain of human sortilin/nts3 in2 complex with neurotensin
51 c2fo0A_
not modelled
6.2
12
PDB header: transferaseChain: A: PDB Molecule: proto-oncogene tyrosine-protein kinase abl1 (1b isoform);PDBTitle: organization of the sh3-sh2 unit in active and inactive forms of the2 c-abl tyrosine kinase
52 c3er0A_
not modelled
6.1
17
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 5a-2;PDBTitle: crystal structure of the full length eif5a from2 saccharomyces cerevisiae
53 c2rf4A_
not modelled
5.6
36
PDB header: transferaseChain: A: PDB Molecule: dna-directed rna polymerase i subunit rpa4;PDBTitle: crystal structure of the rna polymerase i subcomplex a14/43
54 d1umga_
not modelled
5.3
19
Fold: Sulfolobus fructose-1,6-bisphosphatase-likeSuperfamily: Sulfolobus fructose-1,6-bisphosphatase-likeFamily: Sulfolobus fructose-1,6-bisphosphatase-like55 c2htuA_
not modelled
5.2
31
PDB header: hydrolaseChain: A: PDB Molecule: neuraminidase;PDBTitle: n8 neuraminidase in complex with peramivir
56 d1unaa_
not modelled
5.2
39
Fold: RNA bacteriophage capsid proteinSuperfamily: RNA bacteriophage capsid proteinFamily: RNA bacteriophage capsid protein57 c2b8kG_
not modelled
5.1
36
PDB header: transferaseChain: G: PDB Molecule: dna-directed rna polymerase ii 19 kdaPDBTitle: 12-subunit rna polymerase ii