1 c3etnD_
100.0
31
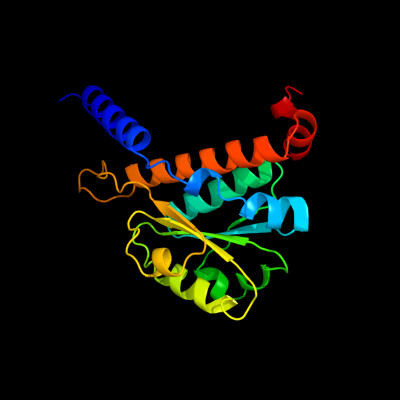
PDB header: isomeraseChain: D: PDB Molecule: putative phosphosugar isomerase involved in capsulePDBTitle: crystal structure of putative phosphosugar isomerase involved in2 capsule formation (yp_209877.1) from bacteroides fragilis nctc 93433 at 1.70 a resolution
2 c3fxaA_
100.0
32
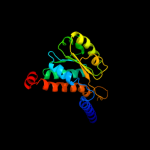
PDB header: sugar binding proteinChain: A: PDB Molecule: sis domain protein;PDBTitle: crystal structure of a putative sugar-phosphate isomerase2 (lmof2365_0531) from listeria monocytogenes str. 4b f2365 at 1.60 a3 resolution
3 c2xhzC_
100.0
49
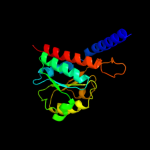
PDB header: isomeraseChain: C: PDB Molecule: arabinose 5-phosphate isomerase;PDBTitle: probing the active site of the sugar isomerase domain from e. coli2 arabinose-5-phosphate isomerase via x-ray crystallography
4 c3cvjB_
100.0
19
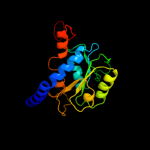
PDB header: isomeraseChain: B: PDB Molecule: putative phosphoheptose isomerase;PDBTitle: crystal structure of a putative phosphoheptose isomerase (bh3325) from2 bacillus halodurans c-125 at 2.00 a resolution
5 d1vima_
100.0
23
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain6 d1m3sa_
100.0
18
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain7 d1jeoa_
99.9
17
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain8 c3shoA_
99.9
15
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, rpir family;PDBTitle: crystal structure of rpir transcription factor from sphaerobacter2 thermophilus (sugar isomerase domain)
9 c2yvaB_
99.9
19
PDB header: dna binding proteinChain: B: PDB Molecule: dnaa initiator-associating protein diaa;PDBTitle: crystal structure of escherichia coli diaa
10 c2puwA_
99.9
10
PDB header: transferaseChain: A: PDB Molecule: isomerase domain of glutamine-fructose-6-phosphatePDBTitle: the crystal structure of isomerase domain of glucosamine-6-phosphate2 synthase from candida albicans
11 d1nria_
99.9
16
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain12 c1nriA_
99.9
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein hi0754;PDBTitle: crystal structure of putative phosphosugar isomerase hi0754 from2 haemophilus influenzae
13 d1x92a_
99.9
18
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain14 c2zj3A_
99.9
13
PDB header: transferaseChain: A: PDB Molecule: glucosamine--fructose-6-phosphatePDBTitle: isomerase domain of human glucose:fructose-6-phosphate2 amidotransferase
15 c3fj1A_
99.9
21
PDB header: isomeraseChain: A: PDB Molecule: putative phosphosugar isomerase;PDBTitle: crystal structure of putative phosphosugar isomerase (yp_167080.1)2 from silicibacter pomeroyi dss-3 at 1.75 a resolution
16 d1tk9a_
99.9
15
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain17 c2amlB_
99.9
13
PDB header: transferaseChain: B: PDB Molecule: sis domain protein;PDBTitle: crystal structure of lmo0035 protein (46906266) from listeria2 monocytogenes 4b f2365 at 1.50 a resolution
18 c3trjC_
99.9
17
PDB header: isomeraseChain: C: PDB Molecule: phosphoheptose isomerase;PDBTitle: structure of a phosphoheptose isomerase from francisella tularensis
19 c2x3yA_
99.9
16
PDB header: isomeraseChain: A: PDB Molecule: phosphoheptose isomerase;PDBTitle: crystal structure of gmha from burkholderia pseudomallei
20 c3hbaA_
99.9
16
PDB header: isomeraseChain: A: PDB Molecule: putative phosphosugar isomerase;PDBTitle: crystal structure of a putative phosphosugar isomerase (sden_2705)2 from shewanella denitrificans os217 at 2.00 a resolution
21 d1j5xa_
not modelled
99.9
18
Fold: SIS domainSuperfamily: SIS domainFamily: double-SIS domain22 d1moqa_
not modelled
99.9
15
Fold: SIS domainSuperfamily: SIS domainFamily: double-SIS domain23 d1x94a_
not modelled
99.9
13
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain24 c3g68A_
not modelled
99.9
13
PDB header: isomeraseChain: A: PDB Molecule: putative phosphosugar isomerase;PDBTitle: crystal structure of a putative phosphosugar isomerase (cd3275) from2 clostridium difficile 630 at 1.80 a resolution
25 c3fnaA_
99.9
36
PDB header: isomeraseChain: A: PDB Molecule: possible arabinose 5-phosphate isomerase;PDBTitle: crystal structure of the cbs pair of possible d-arabinose 5-phosphate2 isomerase yrbh from escherichia coli cft073
26 c3tbfA_
not modelled
99.9
11
PDB header: transferaseChain: A: PDB Molecule: glucosamine--fructose-6-phosphate aminotransferasePDBTitle: c-terminal domain of glucosamine-fructose-6-phosphate aminotransferase2 from francisella tularensis.
27 c3knzA_
not modelled
99.9
15
PDB header: sugar binding proteinChain: A: PDB Molecule: putative sugar binding protein;PDBTitle: crystal structure of putative sugar binding protein (np_459565.1) from2 salmonella typhimurium lt2 at 2.50 a resolution
28 c2a3nA_
not modelled
99.9
18
PDB header: sugar binding proteinChain: A: PDB Molecule: putative glucosamine-fructose-6-phosphate aminotransferase;PDBTitle: crystal structure of a putative glucosamine-fructose-6-phosphate2 aminotransferase (stm4540.s) from salmonella typhimurium lt2 at 1.353 a resolution
29 c1zfjA_
not modelled
99.8
22
PDB header: oxidoreductaseChain: A: PDB Molecule: inosine monophosphate dehydrogenase;PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
30 c3euaD_
not modelled
99.8
16
PDB header: isomeraseChain: D: PDB Molecule: putative fructose-aminoacid-6-phosphate deglycase;PDBTitle: crystal structure of a putative phosphosugar isomerase (bsu32610) from2 bacillus subtilis at 1.90 a resolution
31 c2qh1B_
not modelled
99.8
18
PDB header: unknown functionChain: B: PDB Molecule: hypothetical protein ta0289;PDBTitle: structure of ta289, a cbs-rubredoxin-like protein, in its fe+2-bound2 state
32 c3hf7A_
not modelled
99.8
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized cbs-domain protein;PDBTitle: the crystal structure of a cbs-domain pair with bound amp from2 klebsiella pneumoniae to 2.75a
33 c3pc3A_
not modelled
99.8
15
PDB header: lyaseChain: A: PDB Molecule: cg1753, isoform a;PDBTitle: full length structure of cystathionine beta-synthase from drosophila2 in complex with aminoacrylate
34 c3fkjA_
not modelled
99.8
14
PDB header: isomeraseChain: A: PDB Molecule: putative phosphosugar isomerases;PDBTitle: crystal structure of a putative phosphosugar isomerase (stm_0572) from2 salmonella typhimurium lt2 at 2.12 a resolution
35 d3ddja1
not modelled
99.8
15
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair36 c2emqA_
not modelled
99.8
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical conserved protein;PDBTitle: hypothetical conserved protein (gk1048) from geobacillus kaustophilus
37 c3lhhA_
not modelled
99.8
17
PDB header: membrane proteinChain: A: PDB Molecule: cbs domain protein;PDBTitle: the crystal structure of cbs domain protein from shewanella2 oneidensis mr-1.
38 c1jxaA_
not modelled
99.8
14
PDB header: transferaseChain: A: PDB Molecule: glucosamine 6-phosphate synthase;PDBTitle: glucosamine 6-phosphate synthase with glucose 6-phosphate
39 d1yava3
not modelled
99.8
16
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair40 c2decA_
not modelled
99.8
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 325aa long hypothetical protein;PDBTitle: crystal structure of the ph0510 protein from pyrococcus horikoshii ot3
41 c3i8nB_
not modelled
99.8
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein vp2912;PDBTitle: a domain of a conserved functionally known protein from2 vibrio parahaemolyticus rimd 2210633.
42 c3jtfB_
not modelled
99.8
13
PDB header: transport proteinChain: B: PDB Molecule: magnesium and cobalt efflux protein;PDBTitle: the cbs domain pair structure of a magnesium and cobalt efflux protein2 from bordetella parapertussis in complex with amp
43 c3lfrB_
not modelled
99.8
16
PDB header: transport proteinChain: B: PDB Molecule: putative metal ion transporter;PDBTitle: the crystal structure of a cbs domain from a putative metal2 ion transporter bound to amp from pseudomonas syringae to3 1.55a
44 d1vr9a3
not modelled
99.8
25
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair45 c3lqnA_
not modelled
99.8
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: cbs domain protein;PDBTitle: crystal structure of cbs domain-containing protein of2 unknown function from bacillus anthracis str. ames ancestor
46 c1yavB_
not modelled
99.8
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein bsu14130;PDBTitle: crystal structure of cbs domain-containing protein ykul2 from bacillus subtilis
47 d2yzia1
not modelled
99.8
21
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair48 d1pvma4
not modelled
99.8
18
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair49 c3nqrD_
not modelled
99.8
11
PDB header: transport proteinChain: D: PDB Molecule: magnesium and cobalt efflux protein corc;PDBTitle: a putative cbs domain-containing protein from salmonella typhimurium2 lt2
50 c3ctuB_
not modelled
99.8
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: cbs domain protein;PDBTitle: crystal structure of cbs domain protein from streptococcus2 pneumoniae tigr4
51 c2qlvF_
not modelled
99.8
20
PDB header: transferase/protein bindingChain: F: PDB Molecule: nuclear protein snf4;PDBTitle: crystal structure of the heterotrimer core of the s.2 cerevisiae ampk homolog snf1
52 c1vr9B_
not modelled
99.8
25
PDB header: unknown functionChain: B: PDB Molecule: cbs domain protein/act domain protein;PDBTitle: crystal structure of a cbs domain pair/act domain protein (tm0892)2 from thermotoga maritima at 1.70 a resolution
53 d2nyca1
not modelled
99.8
22
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair54 c2ouxB_
not modelled
99.8
17
PDB header: transport proteinChain: B: PDB Molecule: magnesium transporter;PDBTitle: crystal structure of the soluble part of a magnesium transporter
55 c2qr1E_
not modelled
99.8
19
PDB header: transferaseChain: E: PDB Molecule: protein c1556.08c;PDBTitle: crystal structure of the adenylate sensor from amp-activated protein2 kinase in complex with adp
56 c3ocmA_
not modelled
99.8
16
PDB header: membrane proteinChain: A: PDB Molecule: putative membrane protein;PDBTitle: the crystal structure of a domain from a possible membrane protein of2 bordetella parapertussis
57 d2o16a3
not modelled
99.8
25
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair58 d2yzqa1
not modelled
99.8
20
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair59 c3fwrB_
not modelled
99.8
21
PDB header: transcriptionChain: B: PDB Molecule: yqzb protein;PDBTitle: crystal structure of the cbs domains from the bacillus subtilis ccpn2 repressor complexed with adp
60 c3odpA_
not modelled
99.8
15
PDB header: isomeraseChain: A: PDB Molecule: putative tagatose-6-phosphate ketose/aldose isomerase;PDBTitle: crystal structure of a putative tagatose-6-phosphate ketose/aldose2 isomerase (nt01cx_0292) from clostridium novyi nt at 2.35 a3 resolution
61 d2d4za3
not modelled
99.8
18
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair62 c3lv9A_
not modelled
99.8
18
PDB header: membrane proteinChain: A: PDB Molecule: putative transporter;PDBTitle: crystal structure of cbs domain of a putative transporter from2 clostridium difficile 630
63 d2ef7a1
not modelled
99.8
25
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair64 c3gbyA_
not modelled
99.8
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ct1051;PDBTitle: crystal structure of a protein with unknown function ct10512 from chlorobium tepidum
65 c3orgB_
not modelled
99.8
14
PDB header: transport proteinChain: B: PDB Molecule: cmclc;PDBTitle: crystal structure of a eukaryotic clc transporter
66 c2d4zB_
not modelled
99.8
19
PDB header: transport proteinChain: B: PDB Molecule: chloride channel protein;PDBTitle: crystal structure of the cytoplasmic domain of the chloride channel2 clc-0
67 d1pbja3
not modelled
99.8
20
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair68 d1x9ia_
not modelled
99.7
16
Fold: SIS domainSuperfamily: SIS domainFamily: double-SIS domain69 c2p9mD_
not modelled
99.7
24
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein mj0922;PDBTitle: crystal structure of conserved hypothetical protein mj0922 from2 methanocaldococcus jannaschii dsm 2661
70 d2v8qe2
not modelled
99.7
18
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair71 d1o50a3
not modelled
99.7
15
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair72 d2ouxa2
not modelled
99.7
17
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair73 d1y5ha3
not modelled
99.7
14
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair74 c2v8qE_
not modelled
99.7
17
PDB header: transferaseChain: E: PDB Molecule: 5'-amp-activated protein kinase subunit gamma-1;PDBTitle: crystal structure of the regulatory fragment of mammalian2 ampk in complexes with amp
75 c2yvxD_
not modelled
99.7
24
PDB header: transport proteinChain: D: PDB Molecule: mg2+ transporter mgte;PDBTitle: crystal structure of magnesium transporter mgte
76 c3ocoB_
not modelled
99.7
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hemolysin-like protein containing cbs domains;PDBTitle: the crystal structure of a hemolysin-like protein containing cbs2 domain of oenococcus oeni psu
77 d2j9la1
not modelled
99.7
19
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair78 c2yvzA_
not modelled
99.7
22
PDB header: transport proteinChain: A: PDB Molecule: mg2+ transporter mgte;PDBTitle: crystal structure of magnesium transporter mgte cytosolic domain,2 mg2+-free form
79 c3kpbA_
not modelled
99.7
19
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein mj0100;PDBTitle: crystal structure of the cbs domain pair of protein mj01002 in complex with 5 -methylthioadenosine and s-adenosyl-l-3 methionine.
80 c3kxrA_
not modelled
99.7
18
PDB header: transport proteinChain: A: PDB Molecule: magnesium transporter, putative;PDBTitle: structure of the cystathionine beta-synthase pair domain of the2 putative mg2+ transporter so5017 from shewanella oneidensis mr-1.
81 c3ocmB_
not modelled
99.7
16
PDB header: membrane proteinChain: B: PDB Molecule: putative membrane protein;PDBTitle: the crystal structure of a domain from a possible membrane protein of2 bordetella parapertussis
82 d2ooxe2
not modelled
99.7
20
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair83 d2rc3a1
not modelled
99.7
21
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair84 d2riha1
not modelled
99.7
21
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair85 d2v8qe1
not modelled
99.7
17
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair86 c3l31B_
99.7
30
PDB header: hydrolaseChain: B: PDB Molecule: probable manganase-dependent inorganicPDBTitle: crystal structure of the cbs and drtgg domains of the2 regulatory region of clostridium perfringens3 pyrophosphatase complexed with the inhibitor, amp
87 c3i0zB_
not modelled
99.7
16
PDB header: isomeraseChain: B: PDB Molecule: putative tagatose-6-phosphate ketose/aldose isomerase;PDBTitle: crystal structure of putative putative tagatose-6-phosphate2 ketose/aldose isomerase (np_344614.1) from streptococcus pneumoniae3 tigr4 at 1.70 a resolution
88 c3kh5A_
not modelled
99.7
31
PDB header: unknown functionChain: A: PDB Molecule: protein mj1225;PDBTitle: crystal structure of protein mj1225 from methanocaldococcus2 jannaschii, a putative archaeal homolog of g-ampk.
89 d2ooxe1
not modelled
99.7
13
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair90 c2pfiA_
not modelled
99.7
17
PDB header: transport proteinChain: A: PDB Molecule: chloride channel protein clc-ka;PDBTitle: crystal structure of the cytoplasmic domain of the human2 chloride channel clc-ka
91 c3c3jA_
not modelled
99.7
19
PDB header: isomeraseChain: A: PDB Molecule: putative tagatose-6-phosphate ketose/aldose isomerase;PDBTitle: crystal structure of tagatose-6-phosphate ketose/aldose isomerase from2 escherichia coli
92 c3fhmD_
not modelled
99.7
31
PDB header: structural genomics, unknown function, nChain: D: PDB Molecule: uncharacterized protein atu1752;PDBTitle: crystal structure of the cbs-domain containing protein atu1752 from2 agrobacterium tumefaciens
93 d3ddja2
not modelled
99.7
23
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair94 c3oi8B_
not modelled
99.7
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of functionally unknown conserved protein domain2 from neisseria meningitidis mc58
95 d1zfja4
not modelled
99.6
20
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair96 c3ddjA_
not modelled
99.6
15
PDB header: amp-binding proteinChain: A: PDB Molecule: cbs domain-containing protein;PDBTitle: crystal structure of a cbs domain-containing protein in complex with2 amp (sso3205) from sulfolobus solfataricus at 1.80 a resolution
97 d2yzqa2
not modelled
99.6
14
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair98 c2yzqA_
not modelled
99.6
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative uncharacterized protein ph1780;PDBTitle: crystal structure of uncharacterized conserved protein from2 pyrococcus horikoshii
99 d2yvxa2
not modelled
99.5
24
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair100 c3fioB_
not modelled
99.0
24
PDB header: nucleotide binding protein, metal bindinChain: B: PDB Molecule: a cystathionine beta-synthase domain proteinPDBTitle: crystal structure of a fragment of a cystathionine beta-2 synthase domain protein fused to a zn-ribbon-like domain
101 d1jcna4
not modelled
98.4
18
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair102 c1zzgB_
not modelled
98.2
22
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: crystal structure of hypothetical protein tt0462 from thermus2 thermophilus hb8
103 c2q8nB_
not modelled
98.2
19
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: crystal structure of glucose-6-phosphate isomerase (ec2 5.3.1.9) (tm1385) from thermotoga maritima at 1.82 a3 resolution
104 d1c7qa_
not modelled
98.1
24
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI105 c3ff1B_
not modelled
98.0
23
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: structure of glucose 6-phosphate isomerase from staphylococcus aureus
106 d1jr1a4
not modelled
97.6
25
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair107 d1gzda_
not modelled
97.4
17
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI108 c3jx9B_
not modelled
97.3
12
PDB header: isomeraseChain: B: PDB Molecule: putative phosphoheptose isomerase;PDBTitle: crystal structure of putative phosphoheptose isomerase2 (yp_001815198.1) from exiguobacterium sp. 255-15 at 1.95 a resolution
109 c3ljkA_
not modelled
97.3
22
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: glucose-6-phosphate isomerase from francisella tularensis.
110 d1u0fa_
not modelled
97.2
16
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI111 d1iata_
not modelled
97.2
14
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI112 c2wu8A_
not modelled
97.2
22
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: structural studies of phosphoglucose isomerase from2 mycobacterium tuberculosis h37rv
113 d1q50a_
not modelled
97.1
17
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI114 c3hjbA_
not modelled
97.1
19
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: 1.5 angstrom crystal structure of glucose-6-phosphate isomerase from2 vibrio cholerae.
115 d1hm5a_
not modelled
97.1
18
Fold: SIS domainSuperfamily: SIS domainFamily: Phosphoglucose isomerase, PGI116 c3ujhB_
not modelled
97.1
14
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: crystal structure of substrate-bound glucose-6-phosphate isomerase2 from toxoplasma gondii
117 c3nbuC_
not modelled
97.0
19
PDB header: isomeraseChain: C: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: crystal structure of pgi glucosephosphate isomerase
118 c1t10A_
not modelled
96.8
17
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: phosphoglucose isomerase from leishmania mexicana in complex with2 substrate d-fructose-6-phosphate
119 c2o2cB_
not modelled
96.8
18
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase, glycosomal;PDBTitle: crystal structure of phosphoglucose isomerase from t. brucei2 containing glucose-6-phosphate in the active site
120 c3pr3B_
not modelled
96.8
13
PDB header: isomeraseChain: B: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: crystal structure of plasmodium falciparum glucose-6-phosphate2 isomerase (pf14_0341) in complex with fructose-6-phosphate