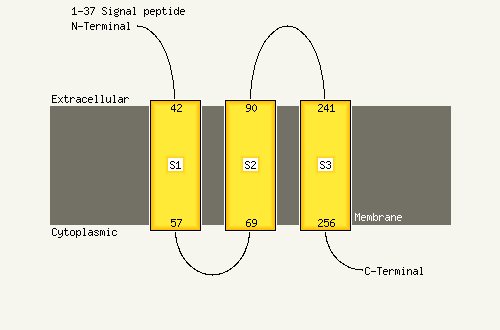
1 c2w8aC_
64.8
11
PDB header: membrane proteinChain: C: PDB Molecule: glycine betaine transporter betp;PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
2 d1jkva_
22.1
15
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Manganese catalase (T-catalase)3 d1go5a_
19.7
8
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TAP-C domain-like4 c2qwuB_
17.8
35
PDB header: cell invasionChain: B: PDB Molecule: intracellular growth locus, subunit c;PDBTitle: crystal structure of f. tularensis pathogenicity island2 protein c
5 d2cwla1
16.4
18
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Manganese catalase (T-catalase)6 d1oaia_
16.2
8
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TAP-C domain-like7 d1joga_
15.3
15
Fold: Four-helical up-and-down bundleSuperfamily: Nucleotidyltransferase substrate binding subunit/domainFamily: Family 1 bi-partite nucleotidyltransferase subunit8 d1wtya_
14.7
18
Fold: Four-helical up-and-down bundleSuperfamily: Nucleotidyltransferase substrate binding subunit/domainFamily: Family 1 bi-partite nucleotidyltransferase subunit9 c3gkxB_
14.3
2
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative arsc family related protein;PDBTitle: crystal structure of putative arsc family related protein from2 bacteroides fragilis
10 c3cuqA_
14.2
30
PDB header: protein transportChain: A: PDB Molecule: vacuolar-sorting protein snf8;PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
11 c2zmeA_
13.5
30
PDB header: protein transportChain: A: PDB Molecule: vacuolar-sorting protein snf8;PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
12 d2gykb1
13.2
23
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: HNH-motif13 d1e8oa_
12.9
10
Fold: Signal recognition particle alu RNA binding heterodimer, SRP9/14Superfamily: Signal recognition particle alu RNA binding heterodimer, SRP9/14Family: Signal recognition particle alu RNA binding heterodimer, SRP9/1414 c2rddB_
12.4
8
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
15 c2jobA_
12.1
30
PDB header: lipid binding proteinChain: A: PDB Molecule: antilipopolysaccharide factor;PDBTitle: solution structure of an antilipopolysaccharide factor from2 shrimp and its possible lipid a binding site
16 c1914A_
12.0
23
PDB header: alu domainChain: A: PDB Molecule: signal recognition particle 9/14 fusion protein;PDBTitle: signal recognition particle alu rna binding heterodimer, srp9/14
17 c1zdbA_
11.9
25
PDB header: igg binding domainChain: A: PDB Molecule: mini protein a domain, z38;PDBTitle: phage-selected mini protein a domain, z38, nmr, minimized2 mean structure
18 c1wwpA_
11.7
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ttha0636;PDBTitle: crystal structure of ttk003001694 from thermus thermophilus2 hb8
19 d2rh2a1
10.6
45
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: R67 dihydrofolate reductase20 d1qqra_
9.0
42
Fold: beta-Grasp (ubiquitin-like)Superfamily: Staphylokinase/streptokinaseFamily: Staphylokinase/streptokinase21 c3fz4A_
not modelled
8.6
2
PDB header: oxidoreductaseChain: A: PDB Molecule: putative arsenate reductase;PDBTitle: the crystal structure of a possible arsenate reductase from2 streptococcus mutans ua159
22 d1dd4d_
not modelled
8.4
27
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain23 d1f6va_
not modelled
8.2
20
Fold: C-terminal domain of B transposition proteinSuperfamily: C-terminal domain of B transposition proteinFamily: C-terminal domain of B transposition protein24 c2vqcA_
not modelled
7.9
17
PDB header: dna-binding proteinChain: A: PDB Molecule: hypothetical 13.2 kda protein;PDBTitle: structure of a dna binding winged-helix protein, f-112,2 from sulfolobus spindle-shaped virus 1.
25 d2vqca1
not modelled
7.9
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: F112-like26 c3kcvG_
not modelled
7.9
13
PDB header: transport proteinChain: G: PDB Molecule: probable formate transporter 1;PDBTitle: structure of formate channel
27 d1eexa_
not modelled
7.6
20
Fold: TIM beta/alpha-barrelSuperfamily: Cobalamin (vitamin B12)-dependent enzymesFamily: Diol dehydratase, alpha subunit28 d1tz7a1
not modelled
7.6
19
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain29 d2p12a1
not modelled
7.5
17
Fold: FomD barrel-likeSuperfamily: FomD-likeFamily: FomD-like30 d2jioa2
not modelled
7.4
13
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-331 c3l78A_
not modelled
7.3
6
PDB header: transcriptionChain: A: PDB Molecule: regulatory protein spx;PDBTitle: the crystal structure of smu.1142c from streptococcus mutans ua159
32 d1eswa_
not modelled
7.2
11
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain33 d1qusa_
not modelled
7.1
15
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: Bacterial muramidase, catalytic domain34 c3sokB_
not modelled
6.9
35
PDB header: cell adhesionChain: B: PDB Molecule: fimbrial protein;PDBTitle: dichelobacter nodosus pilin fima
35 d2o8ia1
not modelled
6.9
21
Fold: UraD-likeSuperfamily: UraD-LikeFamily: UraD-like36 d1914a1
not modelled
6.7
20
Fold: Signal recognition particle alu RNA binding heterodimer, SRP9/14Superfamily: Signal recognition particle alu RNA binding heterodimer, SRP9/14Family: Signal recognition particle alu RNA binding heterodimer, SRP9/1437 d1oqwa_
not modelled
6.2
41
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin38 c2v5iA_
not modelled
6.2
16
PDB header: viral proteinChain: A: PDB Molecule: salmonella typhimurium db7155 bacteriophage det7PDBTitle: structure of the receptor-binding protein of bacteriophage2 det7: a podoviral tailspike in a myovirus
39 c3klzE_
not modelled
6.2
12
PDB header: membrane proteinChain: E: PDB Molecule: putative formate transporter 1;PDBTitle: pentameric formate channel with formate bound
40 c1svfB_
not modelled
6.0
35
PDB header: viral proteinChain: B: PDB Molecule: protein (fusion glycoprotein);PDBTitle: paramyxovirus sv5 fusion protein core
41 d1x1na1
not modelled
6.0
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain42 d2fug34
not modelled
5.7
40
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins43 c2p7vA_
not modelled
5.6
33
PDB header: transcriptionChain: A: PDB Molecule: regulator of sigma d;PDBTitle: crystal structure of the escherichia coli regulator of sigma 70, rsd,2 in complex with sigma 70 domain 4
44 d2pila_
not modelled
5.6
29
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin45 d1pyta_
not modelled
5.4
16
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Pancreatic carboxypeptidase, activation domain46 c1bhbA_
not modelled
5.3
24
PDB header: photoreceptorChain: A: PDB Molecule: bacteriorhodopsin;PDBTitle: three-dimensional structure of (1-71) bacterioopsin2 solubilized in methanol-chloroform and sds micelles3 determined by 15n-1h heteronuclear nmr spectroscopy
47 c2k85A_
not modelled
5.2
21
PDB header: protein bindingChain: A: PDB Molecule: glucocorticoid receptor dna-binding factor 1;PDBTitle: p190-a rhogap ff1 domain
48 c3m8eA_
not modelled
5.1
19
PDB header: dna binding proteinChain: A: PDB Molecule: putative dna-binding protein;PDBTitle: protein structure of type iii plasmid segregation tubr
49 d2q37a1
not modelled
5.1
21
Fold: UraD-likeSuperfamily: UraD-LikeFamily: UraD-like50 c2q37A_
not modelled
5.1
21
PDB header: plant protein, lyaseChain: A: PDB Molecule: ohcu decarboxylase;PDBTitle: crystal structure of ohcu decarboxylase in the presence of2 (s)-allantoin