| 1 |
|
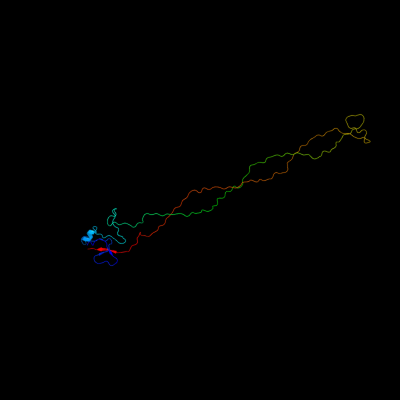
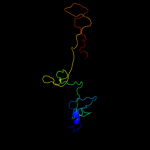
PDB 2xgf chain A
Region: 90 - 320
Aligned: 215
Modelled: 216
Confidence: 100.0%
Identity: 70%
PDB header:viral protein
Chain: A: PDB Molecule:long tail fiber protein p37;
PDBTitle: structure of the bacteriophage t4 long tail fibre needle-2 shaped receptor-binding tip
Phyre2
| 2 |
|
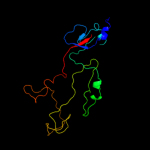
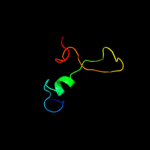
PDB 1pdi chain Q
Region: 81 - 319
Aligned: 186
Modelled: 186
Confidence: 100.0%
Identity: 19%
PDB header:structural protein
Chain: Q: PDB Molecule:short tail fiber protein;
PDBTitle: fitting of the c-terminal part of the short tail fibers2 into the cryo-em reconstruction of t4 baseplate
Phyre2
| 3 |
|
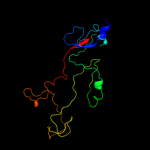
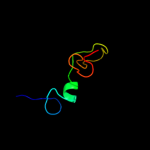
PDB 1ocy chain A
Region: 83 - 319
Aligned: 177
Modelled: 179
Confidence: 100.0%
Identity: 21%
Fold: Receptor-binding domain of short tail fibre protein gp12
Superfamily: Receptor-binding domain of short tail fibre protein gp12
Family: Receptor-binding domain of short tail fibre protein gp12
Phyre2
| 4 |
|
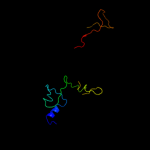
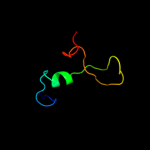
PDB 1h6w chain A
Region: 4 - 137
Aligned: 131
Modelled: 134
Confidence: 99.5%
Identity: 17%
PDB header:structural protein
Chain: A: PDB Molecule:bacteriophage t4 short tail fibre;
PDBTitle: crystal structure of a heat- and protease-stable fragment2 of the bacteriophage t4 short fibre
Phyre2
| 5 |
|
PDB 2fkk chain A
Region: 79 - 200
Aligned: 104
Modelled: 105
Confidence: 88.3%
Identity: 15%
PDB header:viral protein
Chain: A: PDB Molecule:baseplate structural protein gp10;
PDBTitle: crystal structure of the c-terminal domain of the bacteriophage t42 gene product 10
Phyre2
| 6 |
|
PDB 2fl8 chain N
Region: 79 - 213
Aligned: 122
Modelled: 134
Confidence: 68.1%
Identity: 16%
PDB header:virus/viral protein
Chain: N: PDB Molecule:baseplate structural protein gp10;
PDBTitle: fitting of the gp10 trimer structure into the cryoem map of the2 bacteriophage t4 baseplate in the hexagonal conformation.
Phyre2
| 7 |
|
PDB 1m5h chain A domain 2
Region: 94 - 138
Aligned: 39
Modelled: 45
Confidence: 38.2%
Identity: 26%
Fold: Ferredoxin-like
Superfamily: Formylmethanofuran:tetrahydromethanopterin formyltransferase
Family: Formylmethanofuran:tetrahydromethanopterin formyltransferase
Phyre2
| 8 |
|
PDB 1m5s chain A domain 2
Region: 93 - 138
Aligned: 40
Modelled: 46
Confidence: 33.9%
Identity: 23%
Fold: Ferredoxin-like
Superfamily: Formylmethanofuran:tetrahydromethanopterin formyltransferase
Family: Formylmethanofuran:tetrahydromethanopterin formyltransferase
Phyre2
| 9 |
|
PDB 1m5h chain F
Region: 94 - 139
Aligned: 40
Modelled: 46
Confidence: 21.3%
Identity: 25%
PDB header:transferase
Chain: F: PDB Molecule:formylmethanofuran--tetrahydromethanopterin
PDBTitle: formylmethanofuran:tetrahydromethanopterin2 formyltransferase from archaeoglobus fulgidus
Phyre2
| 10 |
|
PDB 1m5s chain C
Region: 94 - 138
Aligned: 39
Modelled: 45
Confidence: 17.6%
Identity: 23%
PDB header:transferase
Chain: C: PDB Molecule:formylmethanofuran--tetrahydromethanopterin
PDBTitle: formylmethanofuran:tetrahydromethanopterin2 fromyltransferase from methanosarcina barkeri
Phyre2
| 11 |
|
PDB 2pfc chain A
Region: 116 - 141
Aligned: 26
Modelled: 26
Confidence: 9.9%
Identity: 38%
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein rv0098/mt0107;
PDBTitle: structure of mycobacterium tuberculosis rv0098
Phyre2
| 12 |
|
PDB 2x3l chain A
Region: 91 - 104
Aligned: 14
Modelled: 14
Confidence: 7.8%
Identity: 43%
PDB header:lyase
Chain: A: PDB Molecule:orn/lys/arg decarboxylase family protein;
PDBTitle: crystal structure of the orn_lys_arg decarboxylase family2 protein sar0482 from methicillin-resistant staphylococcus3 aureus
Phyre2
| 13 |
|
PDB 1fmd chain 1
Region: 91 - 189
Aligned: 58
Modelled: 72
Confidence: 7.4%
Identity: 28%
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily: Positive stranded ssRNA viruses
Family: Picornaviridae-like VP (VP1, VP2, VP3 and VP4)
Phyre2
| 14 |
|
PDB 1ftr chain A domain 2
Region: 94 - 137
Aligned: 38
Modelled: 44
Confidence: 7.2%
Identity: 24%
Fold: Ferredoxin-like
Superfamily: Formylmethanofuran:tetrahydromethanopterin formyltransferase
Family: Formylmethanofuran:tetrahydromethanopterin formyltransferase
Phyre2
| 15 |
|
PDB 1k25 chain A domain 2
Region: 133 - 142
Aligned: 10
Modelled: 10
Confidence: 7.0%
Identity: 60%
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domain
Superfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain
Family: Penicillin-binding protein 2x (pbp-2x), c-terminal domain
Phyre2
| 16 |
|
PDB 1pyy chain A domain 2
Region: 132 - 142
Aligned: 11
Modelled: 11
Confidence: 6.8%
Identity: 55%
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domain
Superfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain
Family: Penicillin-binding protein 2x (pbp-2x), c-terminal domain
Phyre2
| 17 |
|
PDB 1c4k chain A domain 3
Region: 91 - 101
Aligned: 11
Modelled: 11
Confidence: 6.3%
Identity: 36%
Fold: Ornithine decarboxylase C-terminal domain
Superfamily: Ornithine decarboxylase C-terminal domain
Family: Ornithine decarboxylase C-terminal domain
Phyre2
| 18 |
|
PDB 2wzr chain 1
Region: 91 - 130
Aligned: 39
Modelled: 40
Confidence: 5.8%
Identity: 18%
PDB header:virus
Chain: 1: PDB Molecule:polyprotein;
PDBTitle: the structure of foot and mouth disease virus serotype sat1
Phyre2
| 19 |
|
PDB 3kio chain B
Region: 95 - 142
Aligned: 37
Modelled: 48
Confidence: 5.7%
Identity: 19%
PDB header:hydrolase
Chain: B: PDB Molecule:ribonuclease h2 subunit b;
PDBTitle: mouse rnase h2 complex
Phyre2
| 20 |
|
PDB 3n75 chain E
Region: 91 - 101
Aligned: 11
Modelled: 11
Confidence: 5.5%
Identity: 45%
PDB header:lyase
Chain: E: PDB Molecule:lysine decarboxylase, inducible;
PDBTitle: x-ray crystal structure of the escherichia coli inducible lysine2 decarboxylase ldci
Phyre2
| 21 |
|
| 22 |
|