| 1 |
|
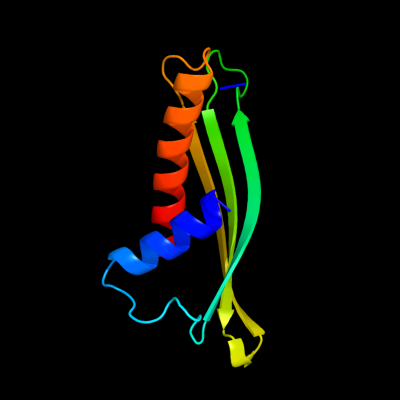
PDB 2iqi chain A domain 1
Region: 93 - 217
Aligned: 103
Modelled: 104
Confidence: 61.4%
Identity: 9%
Fold: Anticodon-binding domain-like
Superfamily: XCC0632-like
Family: XCC0632-like
Phyre2
| 2 |
|
PDB 2dzj chain A
Region: 156 - 209
Aligned: 45
Modelled: 54
Confidence: 49.8%
Identity: 20%
PDB header:sugar binding protein
Chain: A: PDB Molecule:synaptic glycoprotein sc2;
PDBTitle: 2dzj/solution structure of the n-terminal ubiquitin-like2 domain in human synaptic glycoprotein sc2
Phyre2
| 3 |
|
PDB 1pn2 chain A domain 1
Region: 106 - 185
Aligned: 54
Modelled: 54
Confidence: 38.4%
Identity: 4%
Fold: Thioesterase/thiol ester dehydrase-isomerase
Superfamily: Thioesterase/thiol ester dehydrase-isomerase
Family: MaoC-like
Phyre2
| 4 |
|
PDB 3ly8 chain A
Region: 150 - 216
Aligned: 55
Modelled: 67
Confidence: 20.7%
Identity: 9%
PDB header:signaling protein
Chain: A: PDB Molecule:transcriptional activator cadc;
PDBTitle: crystal structure of mutant d471e of the periplasmic domain of cadc
Phyre2
| 5 |
|
PDB 2hqs chain A domain 2
Region: 157 - 214
Aligned: 46
Modelled: 58
Confidence: 15.3%
Identity: 11%
Fold: Anticodon-binding domain-like
Superfamily: TolB, N-terminal domain
Family: TolB, N-terminal domain
Phyre2
| 6 |
|
PDB 3ov5 chain A
Region: 203 - 216
Aligned: 14
Modelled: 14
Confidence: 14.9%
Identity: 29%
PDB header:protein transport
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: atomic structure of the xanthomonas citri virb7 globular domain.
Phyre2
| 7 |
|
PDB 3nqy chain A
Region: 170 - 182
Aligned: 13
Modelled: 13
Confidence: 12.4%
Identity: 31%
PDB header:hydrolase
Chain: A: PDB Molecule:secreted metalloprotease mcp02;
PDBTitle: crystal structure of the autoprocessed complex of vibriolysin mcp-022 with a single point mutation e346a
Phyre2
| 8 |
|
PDB 2axt chain O domain 1
Region: 159 - 186
Aligned: 28
Modelled: 28
Confidence: 9.8%
Identity: 7%
Fold: Transmembrane beta-barrels
Superfamily: OMPA-like
Family: PsbO-like
Phyre2
| 9 |
|
PDB 2bi0 chain A domain 1
Region: 118 - 186
Aligned: 48
Modelled: 49
Confidence: 9.2%
Identity: 19%
Fold: Thioesterase/thiol ester dehydrase-isomerase
Superfamily: Thioesterase/thiol ester dehydrase-isomerase
Family: MaoC-like
Phyre2
| 10 |
|
PDB 2l4w chain A
Region: 203 - 213
Aligned: 11
Modelled: 11
Confidence: 8.5%
Identity: 27%
PDB header:protein transport
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: nmr structure of the xanthomonas virb7
Phyre2
| 11 |
|
PDB 1mww chain A
Region: 83 - 118
Aligned: 36
Modelled: 36
Confidence: 8.0%
Identity: 6%
Fold: Tautomerase/MIF
Superfamily: Tautomerase/MIF
Family: Hypothetical protein HI1388.1
Phyre2
| 12 |
|
PDB 1yiq chain A
Region: 123 - 182
Aligned: 58
Modelled: 60
Confidence: 7.9%
Identity: 19%
PDB header:oxidoreductase
Chain: A: PDB Molecule:quinohemoprotein alcohol dehydrogenase;
PDBTitle: molecular cloning and structural analysis of2 quinohemoprotein alcohol dehydrogenase adhiig from3 pseudomonas putida hk5. compariison to the other4 quinohemoprotein alcohol dehydrogenase adhiib found in the5 same microorganism.
Phyre2
| 13 |
|
PDB 2cy9 chain A domain 1
Region: 157 - 182
Aligned: 26
Modelled: 26
Confidence: 6.6%
Identity: 15%
Fold: Thioesterase/thiol ester dehydrase-isomerase
Superfamily: Thioesterase/thiol ester dehydrase-isomerase
Family: PaaI/YdiI-like
Phyre2
| 14 |
|
PDB 1zto chain A
Region: 19 - 26
Aligned: 8
Modelled: 8
Confidence: 5.6%
Identity: 50%
PDB header:potassium channel
Chain: A: PDB Molecule:potassium channel protein rck4;
PDBTitle: inactivation gate of potassium channel rck4, nmr, 82 structures
Phyre2
| 15 |
|
PDB 1pu1 chain A
Region: 83 - 108
Aligned: 26
Modelled: 26
Confidence: 5.5%
Identity: 19%
Fold: Hypothetical protein MTH677
Superfamily: Hypothetical protein MTH677
Family: Hypothetical protein MTH677
Phyre2
| 16 |
|
PDB 3e9v chain A domain 1
Region: 83 - 118
Aligned: 35
Modelled: 36
Confidence: 5.4%
Identity: 9%
Fold: BTG domain-like
Superfamily: BTG domain-like
Family: BTG domain-like
Phyre2
| 17 |
|
PDB 2rce chain I
Region: 170 - 213
Aligned: 42
Modelled: 44
Confidence: 5.4%
Identity: 10%
PDB header:hydrolase
Chain: I: PDB Molecule:protease degs;
PDBTitle: dfp modified degs delta pdz
Phyre2
| 18 |
|
PDB 1kb0 chain A
Region: 125 - 179
Aligned: 53
Modelled: 55
Confidence: 5.3%
Identity: 15%
PDB header:oxidoreductase
Chain: A: PDB Molecule:quinohemoprotein alcohol dehydrogenase;
PDBTitle: crystal structure of quinohemoprotein alcohol dehydrogenase from2 comamonas testosteroni
Phyre2