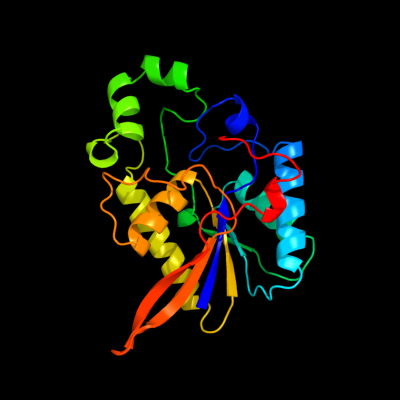
| 1 | d1h2ea_
|
|
 |
100.0 |
31 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Cofactor-dependent phosphoglycerate mutase |
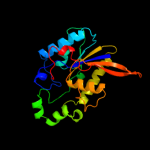
| 2 | d1e58a_
|
|
 |
100.0 |
20 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Cofactor-dependent phosphoglycerate mutase |
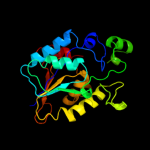
| 3 | c3r7aA_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoglycerate mutase, putative;
PDBTitle: crystal structure of phosphoglycerate mutase from bacillus anthracis2 str. sterne
|
| 4 | c1yjxD_
|
|
 |
100.0 |
22 |
PDB header:isomerase, hydrolase
Chain: D: PDB Molecule:phosphoglycerate mutase 1;
PDBTitle: crystal structure of human b type phosphoglycerate mutase
|
| 5 | c3eznB_
|
|
 |
100.0 |
23 |
PDB header:isomerase
Chain: B: PDB Molecule:2,3-bisphosphoglycerate-dependent phosphoglycerate mutase;
PDBTitle: crystal structure of phosphoglyceromutase from burkholderia2 pseudomallei 1710b
|
| 6 | c2a6pA_
|
|
 |
100.0 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:possible phosphoglycerate mutase gpm2;
PDBTitle: structure solution to 2.2 angstrom and functional characterisation of2 the open reading frame rv3214 from mycobacterium tuberculosis
|
| 7 | c3dcyA_
|
|
 |
100.0 |
31 |
PDB header:apoptosis regulator
Chain: A: PDB Molecule:regulator protein;
PDBTitle: crystal structure a tp53-induced glycolysis and apoptosis2 regulator protein from homo sapiens.
|
| 8 | c3ll4B_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:uncharacterized protein ykr043c;
PDBTitle: structure of the h13a mutant of ykr043c in complex with fructose-1,6-2 bisphosphate
|
| 9 | d1xq9a_
|
|
 |
100.0 |
22 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Cofactor-dependent phosphoglycerate mutase |
| 10 | d1qhfa_
|
|
 |
100.0 |
25 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Cofactor-dependent phosphoglycerate mutase |
| 11 | d1riia_
|
|
 |
100.0 |
24 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Cofactor-dependent phosphoglycerate mutase |
| 12 | c3f3kA_
|
|
 |
100.0 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein ykr043c;
PDBTitle: the structure of uncharacterized protein ykr043c from saccharomyces2 cerevisiae.
|
| 13 | d1fzta_
|
|
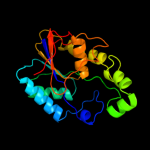 |
100.0 |
23 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Cofactor-dependent phosphoglycerate mutase |
| 14 | d1bifa2
|
|
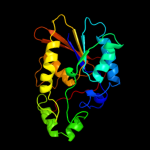 |
100.0 |
22 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, phosphatase domain |
| 15 | c3e9eB_
|
|
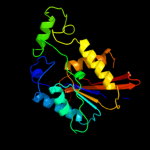 |
100.0 |
31 |
PDB header:hydrolase
Chain: B: PDB Molecule:zgc:56074;
PDBTitle: structure of full-length h11a mutant form of tigar from danio rerio
|
| 16 | d2hhja1
|
|
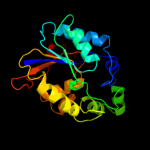 |
100.0 |
21 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Cofactor-dependent phosphoglycerate mutase |
| 17 | c2i1vB_
|
|
 |
100.0 |
22 |
PDB header:transferase, hydrolase
Chain: B: PDB Molecule:6-phosphofructo-2-kinase/fructose-2,6-
PDBTitle: crystal structure of pfkfb3 in complex with adp and2 fructose-2,6-bisphosphate
|
| 18 | c3d8hB_
|
|
 |
100.0 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:glycolytic phosphoglycerate mutase;
PDBTitle: crystal structure of phosphoglycerate mutase from cryptosporidium2 parvum, cgd7_4270
|
| 19 | c1k6mA_
|
|
 |
100.0 |
23 |
PDB header:transferase, hydrolase
Chain: A: PDB Molecule:6-phosphofructo-2-kinase/fructose-2,6-
PDBTitle: crystal structure of human liver 6-phosphofructo-2-2 kinase/fructose-2,6-bisphosphatase
|
| 20 | d3pgma_
|
|
 |
100.0 |
24 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Cofactor-dependent phosphoglycerate mutase |
| 21 | c1bifA_ |
|
not modelled |
100.0 |
23 |
PDB header:bifunctional enzyme
Chain: A: PDB Molecule:6-phosphofructo-2-kinase/ fructose-2,6-bisphosphatase;
PDBTitle: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase bifunctional2 enzyme complexed with atp-g-s and phosphate
|
| 22 | d1k6ma2 |
|
not modelled |
100.0 |
23 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, phosphatase domain |
| 23 | d1tipa_ |
|
not modelled |
100.0 |
22 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase, phosphatase domain |
| 24 | c3hjgB_ |
|
not modelled |
100.0 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative alpha-ribazole-5'-phosphate phosphatase
PDBTitle: crystal structure of putative alpha-ribazole-5'-phosphate2 phosphatase cobc from vibrio parahaemolyticus
|
| 25 | c3d4iD_ |
|
not modelled |
100.0 |
19 |
PDB header:hydrolase
Chain: D: PDB Molecule:sts-2 protein;
PDBTitle: crystal structure of the 2h-phosphatase domain of sts-2
|
| 26 | c2qniA_ |
|
not modelled |
100.0 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu0299;
PDBTitle: crystal structure of uncharacterized protein atu0299
|
| 27 | c2ikqA_ |
|
not modelled |
100.0 |
19 |
PDB header:signaling protein, immune system
Chain: A: PDB Molecule:suppressor of t-cell receptor signaling 1;
PDBTitle: crystal structure of mouse sts-1 pgm domain in complex with phosphate
|
| 28 | c3mxoB_ |
|
not modelled |
100.0 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:serine/threonine-protein phosphatase pgam5, mitochondrial;
PDBTitle: crystal structure oh human phosphoglycerate mutase family member 52 (pgam5)
|
| 29 | d1v37a_ |
|
not modelled |
100.0 |
35 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Cofactor-dependent phosphoglycerate mutase |
| 30 | c3c7tB_ |
|
not modelled |
100.0 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:ecdysteroid-phosphate phosphatase;
PDBTitle: crystal structure of the ecdysone phosphate phosphatase, eppase, from2 bombix mori in complex with tungstate
|
| 31 | c3eozB_ |
|
not modelled |
100.0 |
30 |
PDB header:isomerase
Chain: B: PDB Molecule:putative phosphoglycerate mutase;
PDBTitle: crystal structure of phosphoglycerate mutase from plasmodium2 falciparum, pfd0660w
|
| 32 | c1ujcA_ |
|
not modelled |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphohistidine phosphatase sixa;
PDBTitle: structure of the protein histidine phosphatase sixa2 complexed with tungstate
|
| 33 | c2rflB_ |
|
not modelled |
100.0 |
27 |
PDB header:hydrolase, isomerase
Chain: B: PDB Molecule:putative phosphohistidine phosphatase sixa;
PDBTitle: crystal structure of the putative phosphohistidine phosphatase sixa2 from agrobacterium tumefaciens
|
| 34 | c3f2iD_ |
|
not modelled |
99.9 |
22 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:alr0221 protein;
PDBTitle: crystal structure of the alr0221 protein from nostoc, northeast2 structural genomics consortium target nsr422.
|
| 35 | c3fjyB_ |
|
not modelled |
99.9 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable mutt1 protein;
PDBTitle: crystal structure of a probable mutt1 protein from bifidobacterium2 adolescentis
|
| 36 | c2glcA_ |
|
not modelled |
98.1 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:histidine acid phosphatase;
PDBTitle: structure of francisella tularensis histidine acid2 phosphatase bound to orthovanadate
|
| 37 | d1nd6a_ |
|
not modelled |
98.0 |
22 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Histidine acid phosphatase |
| 38 | d1rpaa_ |
|
not modelled |
97.9 |
25 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Histidine acid phosphatase |
| 39 | d1ihpa_ |
|
not modelled |
97.8 |
22 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Histidine acid phosphatase |
| 40 | d1dkla_ |
|
not modelled |
97.7 |
24 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Histidine acid phosphatase |
| 41 | c2wniC_ |
|
not modelled |
97.7 |
25 |
PDB header:hydrolase
Chain: C: PDB Molecule:3-phytase;
PDBTitle: crystal structure analysis of klebsiella sp asr1 phytase
|
| 42 | d1qwoa_ |
|
not modelled |
97.6 |
17 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Histidine acid phosphatase |
| 43 | d1nt4a_ |
|
not modelled |
97.5 |
16 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Histidine acid phosphatase |
| 44 | d1qfxa_ |
|
not modelled |
97.4 |
26 |
Fold:Phosphoglycerate mutase-like
Superfamily:Phosphoglycerate mutase-like
Family:Histidine acid phosphatase |
| 45 | c2gfiB_ |
|
not modelled |
97.2 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:phytase;
PDBTitle: crystal structure of the phytase from d. castellii at 2.3 a
|
| 46 | c3lfhF_ |
|
not modelled |
35.5 |
25 |
PDB header:transferase
Chain: F: PDB Molecule:phosphotransferase system, mannose/fructose-specific
PDBTitle: crystal structure of manxa from thermoanaerobacter tengcongensis
|
| 47 | d1qgoa_ |
|
not modelled |
34.7 |
15 |
Fold:Chelatase-like
Superfamily:Chelatase
Family:Cobalt chelatase CbiK |
| 48 | d2hk6a1 |
|
not modelled |
33.3 |
15 |
Fold:Chelatase-like
Superfamily:Chelatase
Family:Ferrochelatase |
| 49 | c3mtqA_ |
|
not modelled |
28.7 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:putative phosphoenolpyruvate-dependent sugar
PDBTitle: crystal structure of a putative phosphoenolpyruvate-dependent sugar2 phosphotransferase system (pts) permease (kpn_04802) from klebsiella3 pneumoniae subsp. pneumoniae mgh 78578 at 1.70 a resolution
|
| 50 | c3ngmB_ |
|
not modelled |
26.6 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:extracellular lipase;
PDBTitle: crystal structure of lipase from gibberella zeae
|
| 51 | d1jl3a_ |
|
not modelled |
24.4 |
5 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 52 | d1tiaa_ |
|
not modelled |
24.2 |
22 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Fungal lipases |
| 53 | c3o0dF_ |
|
not modelled |
24.0 |
17 |
PDB header:hydrolase
Chain: F: PDB Molecule:triacylglycerol lipase;
PDBTitle: crystal structure of lip2 lipase from yarrowia lipolytica at 1.7 a2 resolution
|
| 54 | d1dg9a_ |
|
not modelled |
22.6 |
15 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 55 | d1tiba_ |
|
not modelled |
22.3 |
21 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Fungal lipases |
| 56 | c2wmyH_ |
|
not modelled |
19.5 |
15 |
PDB header:hydrolase
Chain: H: PDB Molecule:putative acid phosphatase wzb;
PDBTitle: crystal structure of the tyrosine phosphatase wzb from2 escherichia coli k30 in complex with sulphate.
|
| 57 | c3lp5A_ |
|
not modelled |
19.2 |
6 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative cell surface hydrolase;
PDBTitle: the crystal structure of the putative cell surface hydrolase from2 lactobacillus plantarum wcfs1
|
| 58 | d3tgla_ |
|
not modelled |
18.3 |
17 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Fungal lipases |
| 59 | c2jzcA_ |
|
not modelled |
18.0 |
35 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine transferase subunit
PDBTitle: nmr solution structure of alg13: the sugar donor subunit of2 a yeast n-acetylglucosamine transferase. northeast3 structural genomics consortium target yg1
|
| 60 | d3beda1 |
|
not modelled |
16.8 |
24 |
Fold:PTS system fructose IIA component-like
Superfamily:PTS system fructose IIA component-like
Family:EIIA-man component-like |
| 61 | c2fekA_ |
|
not modelled |
15.8 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight protein-tyrosine-
PDBTitle: structure of a protein tyrosine phosphatase
|
| 62 | c1u2pA_ |
|
not modelled |
15.2 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight protein-tyrosine-
PDBTitle: crystal structure of mycobacterium tuberculosis low2 molecular protein tyrosine phosphatase (mptpa) at 1.9a3 resolution
|
| 63 | d1pdoa_ |
|
not modelled |
15.1 |
29 |
Fold:PTS system fructose IIA component-like
Superfamily:PTS system fructose IIA component-like
Family:EIIA-man component-like |
| 64 | c2o6lA_ |
|
not modelled |
15.1 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:udp-glucuronosyltransferase 2b7;
PDBTitle: crystal structure of the udp-glucuronic acid binding domain2 of the human drug metabolizing udp-glucuronosyltransferase3 2b7
|
| 65 | c3g7nA_ |
|
not modelled |
15.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:lipase;
PDBTitle: crystal structure of a triacylglycerol lipase from2 penicillium expansum at 1.3
|
| 66 | d1l1sa_ |
|
not modelled |
14.9 |
9 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrEF-like |
| 67 | d2ejna1 |
|
not modelled |
14.8 |
29 |
Fold:Uteroglobin-like
Superfamily:Uteroglobin-like
Family:Uteroglobin-like |
| 68 | d1d1qa_ |
|
not modelled |
14.7 |
5 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 69 | d2vcha1 |
|
not modelled |
14.6 |
25 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
| 70 | c3gx1A_ |
|
not modelled |
14.3 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin1832 protein;
PDBTitle: crystal structure of a domain of lin1832 from listeria innocua
|
| 71 | c2gi4A_ |
|
not modelled |
13.9 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:possible phosphotyrosine protein phosphatase;
PDBTitle: solution structure of the low molecular weight protein2 tyrosine phosphatase from campylobacter jejuni.
|
| 72 | d2acva1 |
|
not modelled |
13.3 |
35 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
| 73 | d1vjea_ |
|
not modelled |
13.1 |
13 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 74 | c2l18A_ |
|
not modelled |
12.8 |
5 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:arsenate reductase;
PDBTitle: an arsenate reductase in the phosphate binding state
|
| 75 | c3hbjA_ |
|
not modelled |
12.3 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:flavonoid 3-o-glucosyltransferase;
PDBTitle: structure of ugt78g1 complexed with udp
|
| 76 | d1uwca_ |
|
not modelled |
11.9 |
14 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Fungal lipases |
| 77 | c3iprC_ |
|
not modelled |
11.8 |
12 |
PDB header:transferase
Chain: C: PDB Molecule:pts system, iia component;
PDBTitle: crystal structure of the enterococcus faecalis gluconate2 specific eiia phosphotransferase system component
|
| 78 | c3e90C_ |
|
not modelled |
11.5 |
23 |
PDB header:hydrolase
Chain: C: PDB Molecule:ns2b cofactor;
PDBTitle: west nile vi rus ns2b-ns3protease in complexed with2 inhibitor naph-kkr-h
|
| 79 | c3ds8A_ |
|
not modelled |
11.1 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin2722 protein;
PDBTitle: the crysatl structure of the gene lin2722 products from listeria2 innocua
|
| 80 | d1y1la_ |
|
not modelled |
11.0 |
16 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 81 | d1nvta1 |
|
not modelled |
10.7 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 82 | c2ijoA_ |
|
not modelled |
10.5 |
23 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:polyprotein;
PDBTitle: crystal structure of the west nile virus ns2b-ns3 protease2 complexed with bovine pancreatic trypsin inhibitor
|
| 83 | c2pd2A_ |
|
not modelled |
10.5 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein st0148;
PDBTitle: crystal structure of (st0148) conserved hypothetical from sulfolobus2 tokodaii strain7
|
| 84 | d2pq6a1 |
|
not modelled |
10.0 |
21 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
| 85 | c3lfjB_ |
|
not modelled |
9.9 |
45 |
PDB header:transferase
Chain: B: PDB Molecule:phosphotransferase system, mannose/fructose/n-
PDBTitle: crystal structure of manxb from thermoanaerobacter tengcongensis
|
| 86 | c3jviA_ |
|
not modelled |
9.8 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein tyrosine phosphatase;
PDBTitle: product state mimic crystal structure of protein tyrosine phosphatase2 from entamoeba histolytica
|
| 87 | c2zyiB_ |
|
not modelled |
9.6 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:lipase, putative;
PDBTitle: a. fulgidus lipase with fatty acid fragment and calcium
|
| 88 | d1jf8a_ |
|
not modelled |
9.5 |
9 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 89 | c1zggA_ |
|
not modelled |
9.1 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative low molecular weight protein-tyrosine-
PDBTitle: solution structure of a low molecular weight protein2 tyrosine phosphatase from bacillus subtilis
|
| 90 | d3b48a1 |
|
not modelled |
8.9 |
25 |
Fold:PTS system fructose IIA component-like
Superfamily:PTS system fructose IIA component-like
Family:DhaM-like |
| 91 | d1lgya_ |
|
not modelled |
8.4 |
12 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Fungal lipases |
| 92 | d1lbqa_ |
|
not modelled |
7.3 |
15 |
Fold:Chelatase-like
Superfamily:Chelatase
Family:Ferrochelatase |
| 93 | c3rofA_ |
|
not modelled |
7.3 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight protein-tyrosine-phosphatase ptpa;
PDBTitle: crystal structure of the s. aureus protein tyrosine phosphatase ptpa
|
| 94 | d1nyta1 |
|
not modelled |
7.2 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 95 | d2hrca1 |
|
not modelled |
7.2 |
10 |
Fold:Chelatase-like
Superfamily:Chelatase
Family:Ferrochelatase |
| 96 | c2xvzA_ |
|
not modelled |
7.1 |
14 |
PDB header:metal binding protein
Chain: A: PDB Molecule:chelatase, putative;
PDBTitle: cobalt chelatase cbik (periplasmatic) from desulvobrio2 vulgaris hildenborough (co-crystallized with cobalt)
|
| 97 | c2p10D_ |
|
not modelled |
7.1 |
23 |
PDB header:hydrolase
Chain: D: PDB Molecule:mll9387 protein;
PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
|
| 98 | d2c1xa1 |
|
not modelled |
7.0 |
21 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
| 99 | c3fiuD_ |
|
not modelled |
6.9 |
12 |
PDB header:ligase
Chain: D: PDB Molecule:nh(3)-dependent nad(+) synthetase;
PDBTitle: structure of nmn synthetase from francisella tularensis
|