| 1 |
|
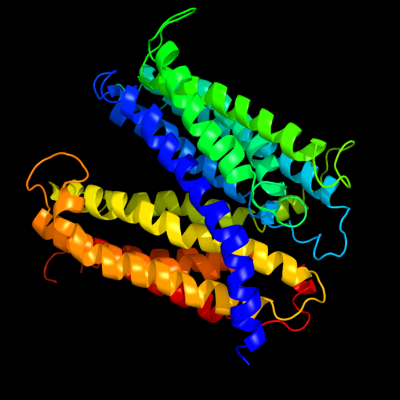
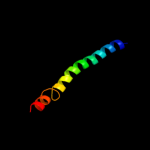
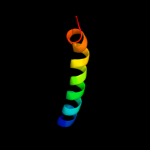
PDB 3mku chain A
Region: 30 - 483
Aligned: 450
Modelled: 450
Confidence: 100.0%
Identity: 20%
PDB header:transport protein
Chain: A: PDB Molecule:multi antimicrobial extrusion protein (na(+)/drug
PDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
Phyre2
| 2 |
|
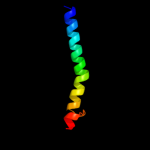
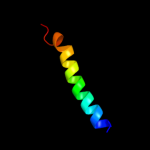
PDB 1pw4 chain A
Region: 23 - 483
Aligned: 421
Modelled: 432
Confidence: 88.8%
Identity: 10%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 3 |
|
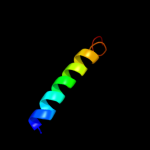
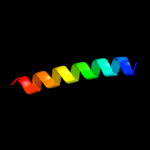
PDB 1pv7 chain A
Region: 40 - 487
Aligned: 411
Modelled: 432
Confidence: 65.7%
Identity: 8%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 4 |
|
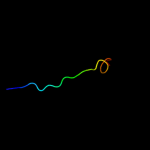
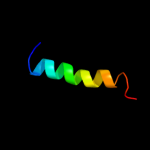
PDB 3qnq chain D
Region: 452 - 490
Aligned: 37
Modelled: 39
Confidence: 36.9%
Identity: 11%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 5 |
|
PDB 2iub chain A domain 2
Region: 419 - 474
Aligned: 47
Modelled: 56
Confidence: 26.7%
Identity: 13%
Fold: Transmembrane helix hairpin
Superfamily: Magnesium transport protein CorA, transmembrane region
Family: Magnesium transport protein CorA, transmembrane region
Phyre2
| 6 |
|
PDB 2knc chain A
Region: 445 - 490
Aligned: 46
Modelled: 46
Confidence: 15.2%
Identity: 15%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 7 |
|
PDB 2p09 chain A
Region: 471 - 493
Aligned: 23
Modelled: 23
Confidence: 12.2%
Identity: 30%
PDB header:de novo protein
Chain: A: PDB Molecule:a non-biological atp binding protein with two mutations
PDBTitle: structural insights into the evolution of a non-biological protein
Phyre2
| 8 |
|
PDB 2oar chain A domain 1
Region: 449 - 490
Aligned: 42
Modelled: 42
Confidence: 11.9%
Identity: 12%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
| 9 |
|
PDB 2knc chain B
Region: 447 - 490
Aligned: 42
Modelled: 44
Confidence: 10.5%
Identity: 12%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 10 |
|
PDB 2oar chain A
Region: 449 - 490
Aligned: 42
Modelled: 42
Confidence: 8.3%
Identity: 12%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2
| 11 |
|
PDB 1zrt chain D
Region: 456 - 482
Aligned: 27
Modelled: 27
Confidence: 6.0%
Identity: 4%
PDB header:oxidoreductase/metal transport
Chain: D: PDB Molecule:cytochrome c1;
PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
Phyre2
| 12 |
|
PDB 2k2o chain A
Region: 471 - 490
Aligned: 20
Modelled: 20
Confidence: 5.7%
Identity: 15%
PDB header:membrane protein
Chain: A: PDB Molecule:myoferlin;
PDBTitle: solution structure of the inner dysf domain of human2 myoferlin
Phyre2
| 13 |
|
PDB 3cwb chain Q
Region: 455 - 482
Aligned: 28
Modelled: 28
Confidence: 5.7%
Identity: 11%
PDB header:oxidoreductase
Chain: Q: PDB Molecule:mitochondrial cytochrome c1, heme protein;
PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
Phyre2
| 14 |
|
PDB 1p84 chain D
Region: 455 - 482
Aligned: 28
Modelled: 28
Confidence: 5.7%
Identity: 11%
PDB header:oxidoreductase
Chain: D: PDB Molecule:cytochrome c1, heme protein;
PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
Phyre2
| 15 |
|
PDB 2yiu chain E
Region: 455 - 477
Aligned: 23
Modelled: 23
Confidence: 5.0%
Identity: 4%
PDB header:oxidoreductase
Chain: E: PDB Molecule:cytochrome c1, heme protein;
PDBTitle: x-ray structure of the dimeric cytochrome bc1 complex from2 the soil bacterium paracoccus denitrificans at 2.73 angstrom resolution
Phyre2
| 16 |
|
PDB 3izs chain Q
Region: 472 - 495
Aligned: 24
Modelled: 24
Confidence: 5.0%
Identity: 25%
PDB header:ribosome
Chain: Q: PDB Molecule:60s ribosomal protein rpl5 (l18p);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
Phyre2