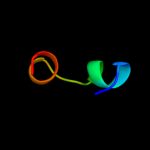| 1 |
|
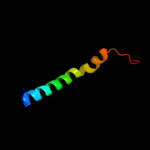
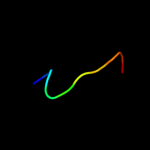
PDB 1w07 chain A domain 2
Region: 32 - 70
Aligned: 39
Modelled: 39
Confidence: 33.1%
Identity: 8%
Fold: Bromodomain-like
Superfamily: Acyl-CoA dehydrogenase C-terminal domain-like
Family: acyl-CoA oxidase C-terminal domains
Phyre2
| 2 |
|
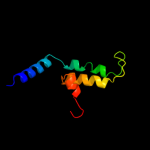
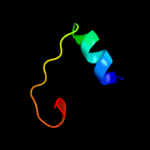
PDB 2ddh chain A domain 2
Region: 32 - 60
Aligned: 29
Modelled: 29
Confidence: 33.0%
Identity: 7%
Fold: Bromodomain-like
Superfamily: Acyl-CoA dehydrogenase C-terminal domain-like
Family: acyl-CoA oxidase C-terminal domains
Phyre2
| 3 |
|
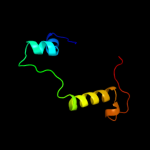
PDB 3iz5 chain K
Region: 73 - 103
Aligned: 27
Modelled: 31
Confidence: 20.2%
Identity: 22%
PDB header:ribosome
Chain: K: PDB Molecule:60s ribosomal protein l13a (l13p);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
Phyre2
| 4 |
|
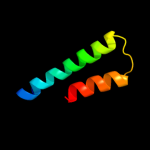
PDB 2ddh chain A
Region: 32 - 70
Aligned: 39
Modelled: 39
Confidence: 15.4%
Identity: 8%
PDB header:oxidoreductase
Chain: A: PDB Molecule:acyl-coa oxidase;
PDBTitle: crystal structure of acyl-coa oxidase complexed with 3-oh-dodecanoate
Phyre2
| 5 |
|
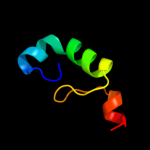
PDB 1w07 chain A
Region: 32 - 70
Aligned: 39
Modelled: 39
Confidence: 14.3%
Identity: 8%
PDB header:oxidoreductase
Chain: A: PDB Molecule:acyl-coa oxidase;
PDBTitle: arabidopsis thaliana acyl-coa oxidase 1
Phyre2
| 6 |
|
PDB 2fon chain A
Region: 32 - 70
Aligned: 39
Modelled: 39
Confidence: 12.8%
Identity: 15%
PDB header:oxidoreductase
Chain: A: PDB Molecule:peroxisomal acyl-coa oxidase 1a;
PDBTitle: x-ray crystal structure of leacx1, an acyl-coa oxidase from2 lycopersicon esculentum (tomato)
Phyre2
| 7 |
|
PDB 3hm5 chain A
Region: 44 - 52
Aligned: 9
Modelled: 9
Confidence: 11.7%
Identity: 44%
PDB header:transcription
Chain: A: PDB Molecule:dna methyltransferase 1-associated protein 1;
PDBTitle: sant domain of human dna methyltransferase 1 associated2 protein 1
Phyre2
| 8 |
|
PDB 4aco chain A
Region: 3 - 76
Aligned: 73
Modelled: 74
Confidence: 9.1%
Identity: 30%
PDB header:dna binding protein
Chain: A: PDB Molecule:centromere dna-binding protein complex cbf3 subunit a;
PDBTitle: structure of the budding yeast ndc10 n-terminal domain
Phyre2
| 9 |
|
PDB 3ag5 chain A
Region: 61 - 106
Aligned: 46
Modelled: 46
Confidence: 9.0%
Identity: 22%
PDB header:ligase
Chain: A: PDB Molecule:pantothenate synthetase;
PDBTitle: crystal structure of pantothenate synthetase from staphylococcus2 aureus
Phyre2
| 10 |
|
PDB 2a6q chain B domain 1
Region: 34 - 72
Aligned: 39
Modelled: 39
Confidence: 8.9%
Identity: 15%
Fold: YefM-like
Superfamily: YefM-like
Family: YefM-like
Phyre2
| 11 |
|
PDB 2js5 chain B
Region: 22 - 41
Aligned: 20
Modelled: 20
Confidence: 8.8%
Identity: 30%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: nmr structure of protein q60c73_metca. northeast structural2 genomics consortium target mcr1
Phyre2
| 12 |
|
PDB 1r5t chain A
Region: 64 - 102
Aligned: 39
Modelled: 39
Confidence: 8.6%
Identity: 13%
Fold: Cytidine deaminase-like
Superfamily: Cytidine deaminase-like
Family: Cytidine deaminase
Phyre2
| 13 |
|
PDB 3las chain A
Region: 62 - 93
Aligned: 31
Modelled: 32
Confidence: 8.5%
Identity: 23%
PDB header:lyase
Chain: A: PDB Molecule:putative carbonic anhydrase;
PDBTitle: crystal structure of carbonic anhydrase from streptococcus mutans to2 1.4 angstrom resolution
Phyre2
| 14 |
|
PDB 4a1a chain I
Region: 73 - 103
Aligned: 27
Modelled: 31
Confidence: 8.5%
Identity: 22%
PDB header:ribosome
Chain: I: PDB Molecule:60s ribosomal protein l13a;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
Phyre2
| 15 |
|
PDB 1iyj chain B domain 2
Region: 66 - 75
Aligned: 10
Modelled: 10
Confidence: 7.4%
Identity: 40%
Fold: BRCA2 tower domain
Superfamily: BRCA2 tower domain
Family: BRCA2 tower domain
Phyre2
| 16 |
|
PDB 1efv chain A domain 2
Region: 83 - 103
Aligned: 21
Modelled: 21
Confidence: 7.3%
Identity: 29%
Fold: DHS-like NAD/FAD-binding domain
Superfamily: DHS-like NAD/FAD-binding domain
Family: C-terminal domain of the electron transfer flavoprotein alpha subunit
Phyre2
| 17 |
|
PDB 2jtv chain A
Region: 47 - 69
Aligned: 23
Modelled: 23
Confidence: 7.1%
Identity: 30%
PDB header:structural genomics
Chain: A: PDB Molecule:protein of unknown function;
PDBTitle: solution structure of protein rpa3401, northeast structural genomics2 consortium target rpt7, ontario center for structural proteomics3 target rp3384
Phyre2
| 18 |
|
PDB 1gve chain A
Region: 34 - 96
Aligned: 43
Modelled: 63
Confidence: 7.1%
Identity: 33%
Fold: TIM beta/alpha-barrel
Superfamily: NAD(P)-linked oxidoreductase
Family: Aldo-keto reductases (NADP)
Phyre2
| 19 |
|
PDB 1v65 chain A
Region: 64 - 79
Aligned: 16
Modelled: 16
Confidence: 7.0%
Identity: 31%
Fold: KRAB domain (Kruppel-associated box)
Superfamily: KRAB domain (Kruppel-associated box)
Family: KRAB domain (Kruppel-associated box)
Phyre2
| 20 |
|
PDB 2fy8 chain A domain 1
Region: 27 - 76
Aligned: 50
Modelled: 50
Confidence: 6.4%
Identity: 16%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: Potassium channel NAD-binding domain
Phyre2
| 21 |
|
| 22 |
|