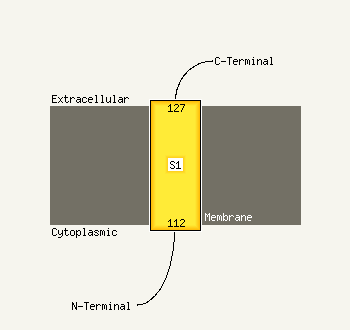
1 d1s4ca1
100.0
28
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: YhcH-like2 d1yfua1
85.4
20
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: 3-hydroxyanthranilic acid dioxygenase-like3 d1v70a_
76.9
17
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: TM1287-like4 c3cewA_
59.9
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized cupin protein;PDBTitle: crystal structure of a cupin protein (bf4112) from bacteroides2 fragilis. northeast structural genomics consortium target bfr205
5 c2i45C_
58.1
22
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of protein nmb1881 from neisseria meningitidis
6 d1zvfa1
41.8
20
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: 3-hydroxyanthranilic acid dioxygenase-like7 d1o4ta_
41.2
24
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: TM1287-like8 d1vj2a_
35.2
16
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: TM1459-like9 d2pa7a1
33.8
13
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: dTDP-sugar isomerase10 d2arca_
33.7
32
Fold: Double-stranded beta-helixSuperfamily: Regulatory protein AraCFamily: Regulatory protein AraC11 c2xxzA_
33.6
21
PDB header: oxidoreductaseChain: A: PDB Molecule: lysine-specific demethylase 6b;PDBTitle: crystal structure of the human jmjd3 jumonji domain
12 d1j3pa_
32.8
24
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Glucose-6-phosphate isomerase, GPI13 c2xueB_
30.6
21
PDB header: oxidoreductaseChain: B: PDB Molecule: lysine-specific demethylase 6b;PDBTitle: crystal structure of jmjd3
14 c3d82A_
30.4
38
PDB header: metal binding proteinChain: A: PDB Molecule: cupin 2, conserved barrel domain protein;PDBTitle: crystal structure of a cupin-2 domain containing protein (sfri_3543)2 from shewanella frigidimarina ncimb 400 at 2.05 a resolution
15 c2ozjB_
27.7
24
PDB header: unknown functionChain: B: PDB Molecule: cupin 2, conserved barrel;PDBTitle: crystal structure of a cupin superfamily protein (dsy2733) from2 desulfitobacterium hafniense dcb-2 at 1.60 a resolution
16 c2fqpD_
27.4
13
PDB header: metal binding proteinChain: D: PDB Molecule: hypothetical protein bp2299;PDBTitle: crystal structure of a cupin domain (bp2299) from bordetella pertussis2 tohama i at 1.80 a resolution
17 c3kgzA_
23.5
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: cupin 2 conserved barrel domain protein;PDBTitle: crystal structure of a cupin 2 conserved barrel domain protein from2 rhodopseudomonas palustris
18 c2oa2A_
22.7
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: bh2720 protein;PDBTitle: crystal structure of bh2720 (10175341) from bacillus halodurans at2 1.41 a resolution
19 d1a6qa1
21.0
16
Fold: Another 3-helical bundleSuperfamily: Protein serine/threonine phosphatase 2C, C-terminal domainFamily: Protein serine/threonine phosphatase 2C, C-terminal domain20 d1y9qa2
15.8
25
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Probable transcriptional regulator VC1968, C-terminal domain21 c2w2iC_
not modelled
14.6
24
PDB header: oxidoreductaseChain: C: PDB Molecule: 2-oxoglutarate oxygenase;PDBTitle: crystal structure of the human 2-oxoglutarate oxygenase2 loc390245
22 c2hdlA_
not modelled
13.6
24
PDB header: cytokineChain: A: PDB Molecule: small inducible cytokine b14;PDBTitle: solution structure of brak/cxcl14
23 d3bb6a1
not modelled
13.1
24
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: TehB-like24 c2os2A_
not modelled
13.1
23
PDB header: oxidoreductaseChain: A: PDB Molecule: jmjc domain-containing histone demethylation protein 3a;PDBTitle: crystal structure of jmjd2a complexed with histone h3 peptide2 trimethylated at lys36
25 c2q8eB_
not modelled
13.0
24
PDB header: oxidoreductaseChain: B: PDB Molecule: jmjc domain-containing histone demethylationPDBTitle: specificity and mechanism of jmjd2a, a trimethyllysine-2 specific histone demethylase
26 c3opwA_
not modelled
13.0
28
PDB header: oxidoreductaseChain: A: PDB Molecule: dna damage-responsive transcriptional repressor rph1;PDBTitle: crystal structure of the rph1 catalytic core
27 d3dl3a1
not modelled
12.9
27
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: TehB-like28 c3ibmB_
not modelled
11.5
23
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: cupin 2, conserved barrel domain protein;PDBTitle: crystal structure of cupin 2 domain-containing protein hhal_0468 from2 halorhodospira halophila
29 c3ebrA_
not modelled
11.2
16
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized rmlc-like cupin;PDBTitle: crystal structure of an rmlc-like cupin protein (reut_a0381) from2 ralstonia eutropha jmp134 at 2.60 a resolution
30 c3fjsC_
not modelled
10.8
27
PDB header: biosynthetic proteinChain: C: PDB Molecule: uncharacterized protein with rmlc-like cupin fold;PDBTitle: crystal structure of a putative biosynthetic protein with rmlc-like2 cupin fold (reut_b4087) from ralstonia eutropha jmp134 at 1.90 a3 resolution
31 d2es7a1
not modelled
10.3
21
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: HyaE-like32 c2pyhB_
not modelled
10.1
26
PDB header: isomeraseChain: B: PDB Molecule: poly(beta-d-mannuronate) c5 epimerase 4;PDBTitle: azotobacter vinelandii mannuronan c-5 epimerase alge4 a-module2 complexed with mannuronan trisaccharide
33 c3h8uA_
not modelled
9.9
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized conserved protein with double-strandedPDBTitle: crystal structure of uncharacterized conserved protein with double-2 stranded beta-helix domain (yp_001338853.1) from klebsiella3 pneumoniae subsp. pneumoniae mgh 78578 at 1.80 a resolution
34 c2r32A_
not modelled
9.8
7
PDB header: immune systemChain: A: PDB Molecule: gcn4-pii/tumor necrosis factor ligandPDBTitle: crystal structure of human gitrl variant
35 c2bx6A_
not modelled
9.6
24
PDB header: transduction proteinChain: A: PDB Molecule: xrp2 protein;PDBTitle: crystal structure of the human retinitis pigmentosa2 protein 2 (rp2)
36 c3l2hD_
not modelled
9.3
18
PDB header: isomeraseChain: D: PDB Molecule: putative sugar phosphate isomerase;PDBTitle: crystal structure of putative sugar phosphate isomerase (afe_0303)2 from acidithiobacillus ferrooxidans atcc 23270 at 1.85 a resolution
37 c2gu9B_
not modelled
9.2
15
PDB header: immune systemChain: B: PDB Molecule: tetracenomycin polyketide synthesis protein;PDBTitle: crystal structure of xc5357 from xanthomonas campestris: a2 putative tetracenomycin polyketide synthesis protein3 adopting a novel cupin subfamily structure
38 c2oziA_
not modelled
8.8
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein rpa4178;PDBTitle: structural genomics, the crystal structure of a putative2 protein rpa4178 from rhodopseudomonas palustris cga009
39 c3ht2A_
not modelled
8.6
28
PDB header: lyaseChain: A: PDB Molecule: remf protein;PDBTitle: zink containing polyketide cyclase remf from streptomyces2 resistomycificus
40 c2q30C_
not modelled
7.7
17
PDB header: unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a rmlc-like cupin protein (dde_2303) from2 desulfovibrio desulfuricans subsp. at 1.94 a resolution
41 c2pfwB_
not modelled
7.6
19
PDB header: unknown functionChain: B: PDB Molecule: cupin 2, conserved barrel domain protein;PDBTitle: crystal structure of a rmlc-like cupin (sfri_3105) from shewanella2 frigidimarina ncimb 400 at 1.90 a resolution
42 c3emrA_
not modelled
7.3
13
PDB header: oxidoreductaseChain: A: PDB Molecule: ectd;PDBTitle: crystal structure analysis of the ectoine hydroxylase ectd from2 salibacillus salexigens
43 d2a1xa1
not modelled
6.6
27
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: PhyH-like44 d1y3ta1
not modelled
6.2
25
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Quercetin 2,3-dioxygenase-like45 d1tvfa1
not modelled
6.2
27
Fold: Penicillin-binding protein associated domainSuperfamily: Penicillin-binding protein associated domainFamily: Pencillin binding protein 4 (PbpD), C-terminal domain46 d2fcta1
not modelled
6.1
18
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: PhyH-like47 c2lazA_
not modelled
5.8
22
PDB header: signaling protein/transcriptionChain: A: PDB Molecule: e3 ubiquitin-protein ligase smurf1;PDBTitle: structure of the first ww domain of human smurf1 in complex with a2 mono-phosphorylated human smad1 derived peptide
48 d1rc6a_
not modelled
5.6
22
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: YlbA-like49 d1j0sa_
not modelled
5.6
24
Fold: beta-TrefoilSuperfamily: CytokineFamily: Interleukin-1 (IL-1)50 c2nnzA_
not modelled
5.5
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: solution structure of the hypothetical protein af2241 from2 archaeoglobus fulgidus
51 c2opwA_
not modelled
5.4
21
PDB header: oxidoreductaseChain: A: PDB Molecule: phyhd1 protein;PDBTitle: crystal structure of human phytanoyl-coa dioxygenase phyhd1 (apo)
52 c2kpsA_
not modelled
5.4
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of domain iv from the ybbr family protein of2 desulfitobacterium hafniense
53 c2lb0A_
not modelled
5.3
22
PDB header: signaling protein/transcriptionChain: A: PDB Molecule: e3 ubiquitin-protein ligase smurf1;PDBTitle: structure of the first ww domain of human smurf1 in complex with a di-2 phosphorylated human smad1 derived peptide