| 1 |
|
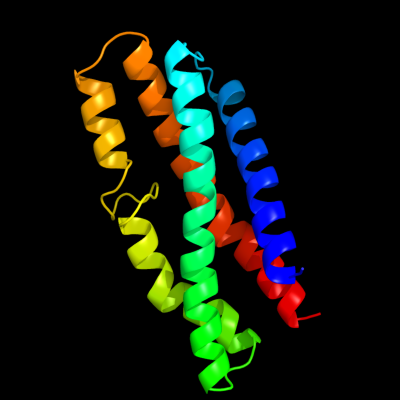
PDB 3aqp chain B
Region: 45 - 184
Aligned: 134
Modelled: 140
Confidence: 54.4%
Identity: 9%
PDB header:membrane protein
Chain: B: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
Phyre2
| 2 |
|
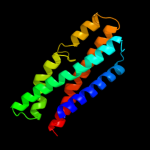
PDB 3k07 chain A
Region: 45 - 191
Aligned: 145
Modelled: 147
Confidence: 51.3%
Identity: 8%
PDB header:transport protein
Chain: A: PDB Molecule:cation efflux system protein cusa;
PDBTitle: crystal structure of cusa
Phyre2
| 3 |
|
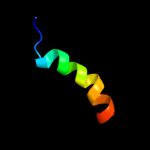
PDB 1oy8 chain A
Region: 45 - 90
Aligned: 46
Modelled: 46
Confidence: 27.7%
Identity: 20%
PDB header:membrane protein
Chain: A: PDB Molecule:acriflavine resistance protein b;
PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
Phyre2
| 4 |
|
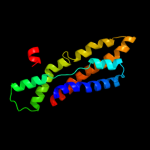
PDB 2vv5 chain D
Region: 6 - 91
Aligned: 86
Modelled: 86
Confidence: 18.3%
Identity: 6%
PDB header:membrane protein
Chain: D: PDB Molecule:small-conductance mechanosensitive channel;
PDBTitle: the open structure of mscs
Phyre2
| 5 |
|
PDB 1hji chain B
Region: 33 - 54
Aligned: 22
Modelled: 22
Confidence: 14.7%
Identity: 18%
PDB header:bacteriophage hk022
Chain: B: PDB Molecule:nun-protein;
PDBTitle: bacteriophage hk022 nun-protein-nutboxb-rna complex
Phyre2
| 6 |
|
PDB 2nn6 chain H domain 3
Region: 33 - 74
Aligned: 42
Modelled: 42
Confidence: 7.6%
Identity: 12%
Fold: Eukaryotic type KH-domain (KH-domain type I)
Superfamily: Eukaryotic type KH-domain (KH-domain type I)
Family: Eukaryotic type KH-domain (KH-domain type I)
Phyre2
| 7 |
|
PDB 2vv5 chain A domain 3
Region: 37 - 91
Aligned: 55
Modelled: 55
Confidence: 6.4%
Identity: 5%
Fold: Mechanosensitive channel protein MscS (YggB), transmembrane region
Superfamily: Mechanosensitive channel protein MscS (YggB), transmembrane region
Family: Mechanosensitive channel protein MscS (YggB), transmembrane region
Phyre2
| 8 |
|
PDB 1iwg chain A domain 8
Region: 45 - 171
Aligned: 124
Modelled: 127
Confidence: 6.1%
Identity: 8%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2