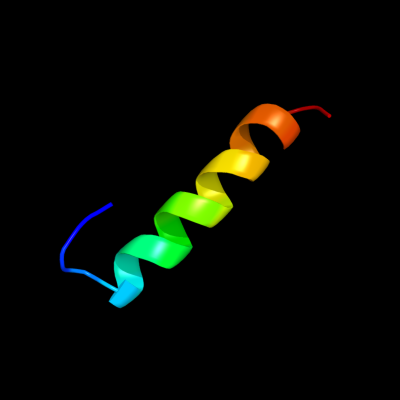
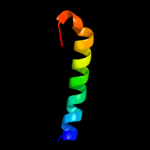
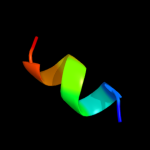
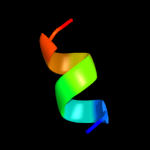
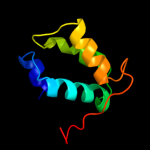
1 c2k19A_
29.8
22
PDB header: antimicrobial proteinChain: A: PDB Molecule: putative piscicolin 126 immunity protein;PDBTitle: nmr solution structure of pisi
2 c3rruA_
27.0
16
PDB header: signaling proteinChain: A: PDB Molecule: tom1l1 protein;PDBTitle: x-ray crystal structure of the vhs domain of human tom1-like protein,2 northeast structural genomics consortium target hr3050e
3 d1mhqa_
26.9
18
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: VHS domain4 d1ujka_
26.1
20
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: VHS domain5 d1wh4a_
24.3
27
Fold: DEATH domainSuperfamily: DEATH domainFamily: DEATH domain, DD6 d1dvpa1
23.7
8
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: VHS domain7 c1iqrA_
23.7
17
PDB header: lyaseChain: A: PDB Molecule: photolyase;PDBTitle: crystal structure of dna photolyase from thermus2 thermophilus
8 d1elka_
21.4
21
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: VHS domain9 c1x5bA_
19.7
25
PDB header: protein bindingChain: A: PDB Molecule: signal transducing adaptor molecule 2;PDBTitle: the solution structure of the vhs domain of human signal2 transducing adaptor molecule 2
10 d1u3da1
18.6
12
Fold: Cryptochrome/photolyase FAD-binding domainSuperfamily: Cryptochrome/photolyase FAD-binding domainFamily: Cryptochrome/photolyase FAD-binding domain11 d1dnpa1
18.2
12
Fold: Cryptochrome/photolyase FAD-binding domainSuperfamily: Cryptochrome/photolyase FAD-binding domainFamily: Cryptochrome/photolyase FAD-binding domain12 c3fefB_
17.1
21
PDB header: hydrolaseChain: B: PDB Molecule: putative glucosidase lpld;PDBTitle: crystal structure of putative glucosidase lpld from2 bacillus subtilis
13 d2bvca2
16.4
43
Fold: Glutamine synthetase/guanido kinaseSuperfamily: Glutamine synthetase/guanido kinaseFamily: Glutamine synthetase catalytic domain14 c3lo0A_
15.5
20
PDB header: hydrolaseChain: A: PDB Molecule: inorganic pyrophosphatase;PDBTitle: crystal structure of inorganic pyrophosphatase from2 ehrlichia chaffeensis
15 c3nj2B_
15.5
6
PDB header: unknown functionChain: B: PDB Molecule: duf269-containing protein;PDBTitle: crystal structure of cce_0566 from the cyanobacterium cyanothece2 51142, a protein associated with nitrogen fixation from the duf2693 family
16 c2jqeA_
15.3
56
PDB header: signaling proteinChain: A: PDB Molecule: signal recognition 54 kda protein;PDBTitle: soution structure of af54 m-domain
17 d2ffha2
15.1
56
Fold: Signal peptide-binding domainSuperfamily: Signal peptide-binding domainFamily: Signal peptide-binding domain18 d1qb2a_
14.8
56
Fold: Signal peptide-binding domainSuperfamily: Signal peptide-binding domainFamily: Signal peptide-binding domain19 c2j4dA_
12.7
16
PDB header: dna-binding proteinChain: A: PDB Molecule: cryptochrome dash;PDBTitle: cryptochrome 3 from arabidopsis thaliana
20 c3fvyA_
12.1
24
PDB header: hydrolaseChain: A: PDB Molecule: dipeptidyl-peptidase 3;PDBTitle: crystal structure of human dipeptidyl peptidase iii
21 d1w7pd1
not modelled
12.1
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain22 d1v66a_
not modelled
12.1
20
Fold: LEM/SAP HeH motifSuperfamily: SAP domainFamily: SAP domain23 d2ebfx1
not modelled
11.5
7
Fold: PMT central region-likeSuperfamily: PMT central region-likeFamily: PMT central region-like24 c2gjhA_
not modelled
11.4
50
PDB header: de novo proteinChain: A: PDB Molecule: designed protein;PDBTitle: nmr structure of cfr (c-terminal fragment of2 computationally designed novel-topology protein top7)
25 c1dvpA_
not modelled
11.3
8
PDB header: transferaseChain: A: PDB Molecule: hepatocyte growth factor-regulated tyrosinePDBTitle: crystal structure of the vhs and fyve tandem domains of hrs,2 a protein involved in membrane trafficking and signal3 transduction
26 c1u3cA_
not modelled
11.1
11
PDB header: signaling proteinChain: A: PDB Molecule: cryptochrome 1 apoprotein;PDBTitle: crystal structure of the phr domain of cryptochrome 1 from2 arabidopsis thaliana
27 d1r76a_
not modelled
11.0
26
Fold: alpha/alpha toroidSuperfamily: Family 10 polysaccharide lyaseFamily: Family 10 polysaccharide lyase28 c1oefA_
not modelled
10.8
33
PDB header: apolipoproteinChain: A: PDB Molecule: apolipoprotein e;PDBTitle: peptide of human apoe residues 263-286, nmr, 5 structures2 at ph 4.8, 37 degrees celsius and peptide:sds mole ratio3 of 1:90
29 d2h80a1
not modelled
10.5
22
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: Variant SAM domain30 d2dkya1
not modelled
10.5
33
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: Variant SAM domain31 c1dnpA_
not modelled
10.4
12
PDB header: lyase (carbon-carbon)Chain: A: PDB Molecule: dna photolyase;PDBTitle: structure of deoxyribodipyrimidine photolyase
32 d1u5tb1
not modelled
10.3
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain33 c2qqoB_
not modelled
10.1
44
PDB header: signaling proteinChain: B: PDB Molecule: neuropilin-2;PDBTitle: crystal structure of the a2b1b2 domains from human neuropilin-2
34 d1vyva2
not modelled
9.8
37
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases35 d1qzxa2
not modelled
9.3
50
Fold: Signal peptide-binding domainSuperfamily: Signal peptide-binding domainFamily: Signal peptide-binding domain36 d2sh1a_
not modelled
9.0
42
Fold: Defensin-likeSuperfamily: Defensin-likeFamily: Defensin37 c3s90D_
not modelled
8.9
55
PDB header: cell adhesionChain: D: PDB Molecule: talin-1;PDBTitle: human vinculin head domain vh1 (residues 1-252) in complex with murine2 talin (vbs33; residues 1512-1546)
38 c1htoB_
not modelled
8.6
43
PDB header: ligaseChain: B: PDB Molecule: glutamine synthetase;PDBTitle: crystallographic structure of a relaxed glutamine synthetase from2 mycobacterium tuberculosis
39 c3g7pA_
not modelled
8.4
17
PDB header: unknown functionChain: A: PDB Molecule: nitrogen fixation protein;PDBTitle: crystal structure of a nifx-associated protein of unknown function2 (afe_1514) from acidithiobacillus ferrooxidans atcc at 2.00 a3 resolution
40 d1f52a2
not modelled
8.2
33
Fold: Glutamine synthetase/guanido kinaseSuperfamily: Glutamine synthetase/guanido kinaseFamily: Glutamine synthetase catalytic domain41 c1s6yA_
not modelled
8.1
19
PDB header: hydrolaseChain: A: PDB Molecule: 6-phospho-beta-glucosidase;PDBTitle: 2.3a crystal structure of phospho-beta-glucosidase
42 c2qqmA_
not modelled
8.1
29
PDB header: signaling proteinChain: A: PDB Molecule: neuropilin-1;PDBTitle: crystal structure of the a2b1b2 domains from human neuropilin-1
43 c2jxfA_
not modelled
8.0
27
PDB header: viral protein, membrane proteinChain: A: PDB Molecule: genome polyprotein;PDBTitle: the solution structure of hcv ns4b(40-69)
44 c1np7A_
not modelled
7.9
14
PDB header: lyaseChain: A: PDB Molecule: dna photolyase;PDBTitle: crystal structure analysis of synechocystis sp. pcc6803 cryptochrome
45 c1u8xX_
not modelled
7.8
15
PDB header: hydrolaseChain: X: PDB Molecule: maltose-6'-phosphate glucosidase;PDBTitle: crystal structure of glva from bacillus subtilis, a metal-requiring,2 nad-dependent 6-phospho-alpha-glucosidase
46 c2iunD_
not modelled
7.8
30
PDB header: viral proteinChain: D: PDB Molecule: avian adenovirus celo long fibre;PDBTitle: structure of the c-terminal head domain of the avian2 adenovirus celo long fibre (p21 crystal form)
47 c3c8zB_
not modelled
7.7
23
PDB header: ligaseChain: B: PDB Molecule: cysteinyl-trna synthetase;PDBTitle: the 1.6 a crystal structure of mshc: the rate limiting2 enzyme in the mycothiol biosynthetic pathway
48 c1fpyE_
not modelled
7.5
33
PDB header: ligaseChain: E: PDB Molecule: glutamine synthetase;PDBTitle: crystal structure of glutamine synthetase from salmonella2 typhimurium with inhibitor phosphinothricin
49 c2hfzA_
not modelled
7.4
16
PDB header: transferaseChain: A: PDB Molecule: rna-directed rna polymerase(ns5);PDBTitle: crystal structure of rna dependent rna polymerase domain2 from west nile virus
50 c1zzpA_
not modelled
7.4
11
PDB header: transferaseChain: A: PDB Molecule: proto-oncogene tyrosine-protein kinase abl1;PDBTitle: solution structure of the f-actin binding domain of bcr-2 abl/c-abl
51 c1vjtA_
not modelled
7.4
12
PDB header: hydrolaseChain: A: PDB Molecule: alpha-glucosidase;PDBTitle: crystal structure of alpha-glucosidase (tm0752) from thermotoga2 maritima at 2.50 a resolution
52 d1np7a1
not modelled
7.1
14
Fold: Cryptochrome/photolyase FAD-binding domainSuperfamily: Cryptochrome/photolyase FAD-binding domainFamily: Cryptochrome/photolyase FAD-binding domain53 c2eodA_
not modelled
7.0
38
PDB header: signaling proteinChain: A: PDB Molecule: tnf receptor-associated factor 4;PDBTitle: solution structure of traf-type zinc finger domains (190-2 248) from human tnf receptor-associated factor 4
54 c3zxpC_
not modelled
6.9
15
PDB header: protein transportChain: C: PDB Molecule: bro1 domain-containing protein brox;PDBTitle: structural and functional analyses of the bro1 domain protein brox
55 d1enwa_
not modelled
6.8
10
Fold: RuvA C-terminal domain-likeSuperfamily: Elongation factor TFIIS domain 2Family: Elongation factor TFIIS domain 256 c3r9mA_
not modelled
6.8
15
PDB header: protein bindingChain: A: PDB Molecule: bro1 domain-containing protein brox;PDBTitle: crystal structure of the brox bro1 domain
57 c1tezB_
not modelled
6.8
11
PDB header: lyase/dnaChain: B: PDB Molecule: deoxyribodipyrimidine photolyase;PDBTitle: complex between dna and the dna photolyase from anacystis nidulans
58 c1oegA_
not modelled
6.7
37
PDB header: apolipoproteinChain: A: PDB Molecule: apolipoprotein e;PDBTitle: peptide of human apoe residues 267-289, nmr, 5 structures2 at ph 6.0, 37 degrees celsius and peptide:sds mole ratio3 of 1:90
59 d1t3la2
not modelled
6.6
53
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases60 d1t0hb_
not modelled
6.5
47
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases61 d1vyua2
not modelled
6.5
47
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases62 d1aq0a_
not modelled
6.4
33
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases63 c3juaB_
not modelled
6.3
16
PDB header: transcriptionChain: B: PDB Molecule: 65 kda yes-associated protein;PDBTitle: structural basis of yap recognition by tead4 in the hippo pathway
64 c2entA_
not modelled
6.0
35
PDB header: transcriptionChain: A: PDB Molecule: krueppel-like factor 15;PDBTitle: solution structure of the second c2h2-type zinc finger2 domain from human krueppel-like factor 15
65 d1bvp11
not modelled
5.9
23
Fold: A virus capsid protein alpha-helical domainSuperfamily: A virus capsid protein alpha-helical domainFamily: Orbivirus capsid66 c1up6F_
not modelled
5.9
22
PDB header: hydrolaseChain: F: PDB Molecule: 6-phospho-beta-glucosidase;PDBTitle: structure of the 6-phospho-beta glucosidase from thermotoga2 maritima at 2.55 angstrom resolution in the tetragonal form3 with manganese, nad+ and glucose-6-phosphate
67 d1oqva_
not modelled
5.7
31
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: TcpA-like pilin68 d2bo9b1
not modelled
5.6
17
Fold: Cystatin-likeSuperfamily: Cystatin/monellinFamily: Latexin-like69 c3mkqB_
not modelled
5.6
12
PDB header: transport proteinChain: B: PDB Molecule: coatomer subunit alpha;PDBTitle: crystal structure of yeast alpha/betaprime-cop subcomplex of the copi2 vesicular coat
70 c2kk1A_
not modelled
5.4
21
PDB header: transferaseChain: A: PDB Molecule: tyrosine-protein kinase abl2;PDBTitle: solution structure of c-terminal domain of tyrosine-protein2 kinase abl2 from homo sapiens, northeast structural3 genomics consortium (nesg) target hr5537a
71 c3cskA_
not modelled
5.2
25
PDB header: hydrolaseChain: A: PDB Molecule: probable dipeptidyl-peptidase 3;PDBTitle: structure of dpp iii from saccharomyces cerevisiae
72 d1hn6a_
not modelled
5.2
30
Fold: Apical membrane antigen 1Superfamily: Apical membrane antigen 1Family: Apical membrane antigen 173 c2v0xB_
not modelled
5.2
21
PDB header: cell cycleChain: B: PDB Molecule: lamina-associated polypeptide 2 isoformsPDBTitle: the dimerization domain of lap2alpha
74 c3fkyD_
not modelled
5.2
14
PDB header: ligaseChain: D: PDB Molecule: glutamine synthetase;PDBTitle: crystal structure of the glutamine synthetase gln1deltan182 from the yeast saccharomyces cerevisiae
75 c2w2uA_
not modelled
5.1
27
PDB header: hydrolase/transportChain: A: PDB Molecule: hypothetical p60 katanin;PDBTitle: structural insight into the interaction between archaeal2 escrt-iii and aaa-atpase
76 d1czsa_
not modelled
5.0
42
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Discoidin domain (FA58C, coagulation factor 5/8 C-terminal domain)77 c2l5rA_
not modelled
5.0
50
PDB header: antimicrobial proteinChain: A: PDB Molecule: antimicrobial peptide alyteserin-1c;PDBTitle: conformational and membrane interactins studies of antimicrobial2 peptide alyteserin-1c