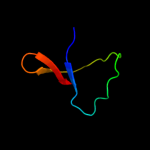
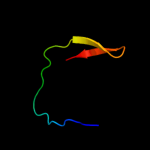
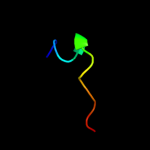
1 c2jsoA_
96.9
19
PDB header: signaling proteinChain: A: PDB Molecule: polymyxin resistance protein pmrd;PDBTitle: antimicrobial resistance protein
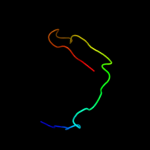
2 c2rqxA_
96.7
17
PDB header: signaling proteinChain: A: PDB Molecule: polymyxin b resistance protein;PDBTitle: solution nmr structure of pmrd from klebsiella pneumoniae
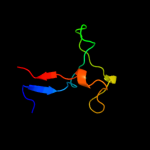
3 c2k75A_
59.1
7
PDB header: dna binding proteinChain: A: PDB Molecule: uncharacterized protein ta0387;PDBTitle: solution nmr structure of the ob domain of ta0387 from2 thermoplasma acidophilum. northeast structural genomics3 consortium target tar80b.
4 c2kenA_
35.7
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein;PDBTitle: solution nmr structure of the ob domain (residues 67-166)2 of mm0293 from methanosarcina mazei. northeast structural3 genomics consortium target mar214a.
5 d1giqa2
27.9
17
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: ADP-ribosylating toxins6 d1r45a_
25.3
21
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: ADP-ribosylating toxins7 c3bw8B_
25.0
28
PDB header: transferaseChain: B: PDB Molecule: mono-adp-ribosyltransferase c3;PDBTitle: crystal structure of the clostridium limosum c3 exoenzyme
8 c1xzzA_
24.7
22
PDB header: membrane proteinChain: A: PDB Molecule: inositol 1,4,5-trisphosphate receptor type 1;PDBTitle: crystal structure of the ligand binding suppressor domain of type 12 inositol 1,4,5-trisphosphate receptor
9 c3dm3A_
23.7
11
PDB header: replicationChain: A: PDB Molecule: replication factor a;PDBTitle: crystal structure of a domain of a replication factor a2 protein, from methanocaldococcus jannaschii. northeast3 structural genomics target mjr118e
10 d1y0jb1
20.5
38
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: C2HC finger11 d1o7ia_
18.8
10
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB12 c2k9oA_
18.2
36
PDB header: toxinChain: A: PDB Molecule: vm24 scorpion toxin;PDBTitle: solution structure of vm24 synthetic scorpion toxin
13 d1wdia_
18.1
38
Fold: QueA-likeSuperfamily: QueA-likeFamily: QueA-like14 c2b68A_
17.6
36
PDB header: antimicrobial proteinChain: A: PDB Molecule: defensin;PDBTitle: solution structure of the recombinant crassostrea gigas2 defensin
15 d1giqa1
17.2
17
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: ADP-ribosylating toxins16 c1hvwA_
16.2
30
PDB header: toxinChain: A: PDB Molecule: omega-atracotoxin-hv1a;PDBTitle: hairpinless mutant of omega-atracotoxin-hv1a
17 d1hvwa_
16.2
30
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins18 c2k6uA_
16.0
57
PDB header: hormoneChain: A: PDB Molecule: insulin-like 3 a chain;PDBTitle: the solution structure of a conformationally restricted2 fully active derivative of the human relaxin-like factor3 (rlf)
19 d2apja1
15.9
40
Fold: Flavodoxin-likeSuperfamily: SGNH hydrolaseFamily: Putative acetylxylan esterase-like20 c1i4oC_
15.1
12
PDB header: apoptosis/hydrolaseChain: C: PDB Molecule: baculoviral iap repeat-containing protein 4;PDBTitle: crystal structure of the xiap/caspase-7 complex
21 d1vkya_
not modelled
14.6
31
Fold: QueA-likeSuperfamily: QueA-likeFamily: QueA-like22 d1bx7a_
not modelled
14.0
42
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Leech antihemostatic proteinsFamily: Huristasin-like23 d1yq2a5
not modelled
13.8
6
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases24 d1zmba1
not modelled
13.5
30
Fold: Flavodoxin-likeSuperfamily: SGNH hydrolaseFamily: Putative acetylxylan esterase-like25 c3u0jB_
not modelled
13.3
21
PDB header: transferaseChain: B: PDB Molecule: type iii effector hopu1;PDBTitle: crystal structure of adp-ribosyltransferase hopu1 of pseudomonas2 syringae pv. tomato dc3000
26 d1fmta1
not modelled
13.0
25
Fold: FMT C-terminal domain-likeSuperfamily: FMT C-terminal domain-likeFamily: Post formyltransferase domain27 c1yy3A_
not modelled
12.8
13
PDB header: isomeraseChain: A: PDB Molecule: s-adenosylmethionine:trna ribosyltransferase-PDBTitle: structure of s-adenosylmethionine:trna ribosyltransferase-2 isomerase (quea)
28 d1rm6c2
not modelled
12.1
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins29 d1skza1
not modelled
12.1
38
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Leech antihemostatic proteinsFamily: Huristasin-like30 d1iray2
not modelled
11.4
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains31 d1l7la_
not modelled
11.3
40
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: PA-IL, galactose-binding lectin 132 c2gwmA_
not modelled
10.9
13
PDB header: transferase, toxinChain: A: PDB Molecule: 65 kda virulence protein;PDBTitle: crystal structure of the salmonella spvb atr domain
33 d1qs1a1
not modelled
10.9
13
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: ADP-ribosylating toxins34 d1ejab_
not modelled
10.4
31
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Leech antihemostatic proteinsFamily: Huristasin-like35 d1hmja_
not modelled
10.2
8
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB536 c2yuzA_
not modelled
10.0
11
PDB header: immune systemChain: A: PDB Molecule: myosin-binding protein c, slow-type;PDBTitle: solution structure of 4th immunoglobulin domain of slow2 type myosin-binding protein c
37 c2bovB_
not modelled
9.9
24
PDB header: transferaseChain: B: PDB Molecule: mono-adp-ribosyltransferase c3;PDBTitle: molecular recognition of an adp-ribosylating clostridium2 botulinum c3 exoenzyme by rala gtpase
38 d1dzfa2
not modelled
9.8
15
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB539 d1eika_
not modelled
9.3
15
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB540 d1wjja_
not modelled
9.2
13
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB41 c3gabC_
not modelled
8.7
50
PDB header: hydrolaseChain: C: PDB Molecule: dna mismatch repair protein mutl;PDBTitle: c-terminal domain of bacillus subtilis mutl crystal form i
42 c1z0zC_
not modelled
8.6
14
PDB header: transferaseChain: C: PDB Molecule: probable inorganic polyphosphate/atp-nad kinase;PDBTitle: crystal structure of a nad kinase from archaeoglobus2 fulgidus in complex with nad
43 d1z0sa1
not modelled
8.6
14
Fold: NAD kinase/diacylglycerol kinase-likeSuperfamily: NAD kinase/diacylglycerol kinase-likeFamily: NAD kinase-like44 c2h6oA_
not modelled
8.6
26
PDB header: viral proteinChain: A: PDB Molecule: major outer envelope glycoprotein gp350;PDBTitle: epstein barr virus major envelope glycoprotein
45 d1axha_
not modelled
8.5
27
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins46 c3pt5A_
not modelled
8.4
20
PDB header: hydrolaseChain: A: PDB Molecule: nans (yjhs), a 9-o-acetyl n-acetylneuraminic acid esterase;PDBTitle: crystal structure of nans
47 d1ojqa_
not modelled
8.3
27
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: ADP-ribosylating toxins48 c2cpcA_
not modelled
8.2
6
PDB header: immune systemChain: A: PDB Molecule: kiaa0657 protein;PDBTitle: solution structure of rsgi ruh-030, an ig like domain from2 human cdna
49 c3bg4D_
not modelled
8.1
38
PDB header: hydrolase/hydrolase inhibitorChain: D: PDB Molecule: guamerin;PDBTitle: the crystal structure of guamerin in complex with2 chymotrypsin and the development of an elastase-specific3 inhibitor
50 d1tdha3
not modelled
7.9
43
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins51 c1wc6B_
not modelled
7.9
28
PDB header: lyaseChain: B: PDB Molecule: adenylate cyclase;PDBTitle: soluble adenylyl cyclase cyac from s. platensis in complex2 with rp-atpalphas in presence of bicarbonate
52 d2c8aa1
not modelled
7.8
24
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: ADP-ribosylating toxins53 c2kqdA_
not modelled
7.8
50
PDB header: lyaseChain: A: PDB Molecule: aprataxin and pnk-like factor;PDBTitle: first pbz domain of human aplf protein in complex with2 ribofuranosyladenosine
54 d2yt9a1
not modelled
7.6
50
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H255 d1uzva_
not modelled
7.6
15
Fold: Calcium-mediated lectinSuperfamily: Calcium-mediated lectinFamily: Calcium-mediated lectin56 d1xiwb_
not modelled
7.6
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains57 d1pcfa_
not modelled
7.6
31
Fold: ssDNA-binding transcriptional regulator domainSuperfamily: ssDNA-binding transcriptional regulator domainFamily: Transcriptional coactivator PC4 C-terminal domain58 c2eo1A_
not modelled
7.4
11
PDB header: contractile proteinChain: A: PDB Molecule: cdna flj14124 fis, clone mamma1002498;PDBTitle: solution structure of the ig domain of human obscn protein
59 d1m0da_
not modelled
7.3
31
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Endonuclease I (Holliday junction resolvase)60 c2pmzV_
not modelled
7.2
23
PDB header: translation, transferaseChain: V: PDB Molecule: dna-directed rna polymerase subunit h;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
61 c3ncvB_
not modelled
7.0
33
PDB header: hydrolaseChain: B: PDB Molecule: dna mismatch repair protein mutl;PDBTitle: ngol
62 c2zfnA_
not modelled
7.0
28
PDB header: transferaseChain: A: PDB Molecule: regulator of ty1 transposition protein 109;PDBTitle: self-acetylation mediated histone h3 lysine 56 acetylation by rtt109
63 d1qs1a2
not modelled
6.9
13
Fold: ADP-ribosylationSuperfamily: ADP-ribosylationFamily: ADP-ribosylating toxins64 c2jtdA_
not modelled
6.9
3
PDB header: cell adhesionChain: A: PDB Molecule: myomesin-1;PDBTitle: skelemin immunoglobulin c2 like domain 4
65 c3mr7B_
not modelled
6.7
24
PDB header: hydrolaseChain: B: PDB Molecule: adenylate/guanylate cyclase/hydrolase, alpha/beta foldPDBTitle: crystal structure of adenylate/guanylate cyclase/hydrolase from2 silicibacter pomeroyi
66 d1usra_
not modelled
6.6
11
Fold: 6-bladed beta-propellerSuperfamily: SialidasesFamily: Sialidases (neuraminidases)67 c3ehkC_
not modelled
6.5
12
PDB header: plant proteinChain: C: PDB Molecule: prunin;PDBTitle: crystal structure of pru du amandin, an allergenic protein2 from prunus dulcis
68 c1rwuA_
not modelled
6.5
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical upf0250 protein ybed;PDBTitle: solution structure of conserved protein ybed from e. coli
69 d1rwua_
not modelled
6.5
13
Fold: Ferredoxin-likeSuperfamily: YbeD/HP0495-likeFamily: YbeD-like70 d1rgoa2
not modelled
6.5
57
Fold: CCCH zinc fingerSuperfamily: CCCH zinc fingerFamily: CCCH zinc finger71 d1wwbx_
not modelled
6.3
7
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains72 d1wd2a_
not modelled
6.2
30
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: RING finger domain, C3HC473 d2nqra2
not modelled
6.0
13
Fold: MoeA N-terminal region -likeSuperfamily: MoeA N-terminal region -likeFamily: MoeA N-terminal region -like74 d1jz8a5
not modelled
5.8
9
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases75 d2isya2
not modelled
5.7
33
Fold: Iron-dependent repressor protein, dimerization domainSuperfamily: Iron-dependent repressor protein, dimerization domainFamily: Iron-dependent repressor protein, dimerization domain76 c2k50A_
not modelled
5.7
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: replication factor a related protein;PDBTitle: solution nmr structure of the replication factor a related2 protein from methanobacterium thermoautotrophicum.3 northeast structural genomics target tr91a.
77 d6paxa1
not modelled
5.5
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain78 c2zkqi_
not modelled
5.5
11
PDB header: ribosomal protein/rnaChain: I: PDB Molecule: PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
79 d2cu3a1
not modelled
5.4
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS80 c2j58G_
not modelled
5.2
25
PDB header: membrane proteinChain: G: PDB Molecule: outer membrane lipoprotein wza;PDBTitle: the structure of wza
81 d1s04a_
not modelled
5.2
16
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain82 c1ti2F_
not modelled
5.2
67
PDB header: oxidoreductaseChain: F: PDB Molecule: pyrogallol hydroxytransferase small subunit;PDBTitle: crystal structure of pyrogallol-phloroglucinol2 transhydroxylase from pelobacter acidigallici
83 d1g3wa2
not modelled
5.1
29
Fold: Iron-dependent repressor protein, dimerization domainSuperfamily: Iron-dependent repressor protein, dimerization domainFamily: Iron-dependent repressor protein, dimerization domain