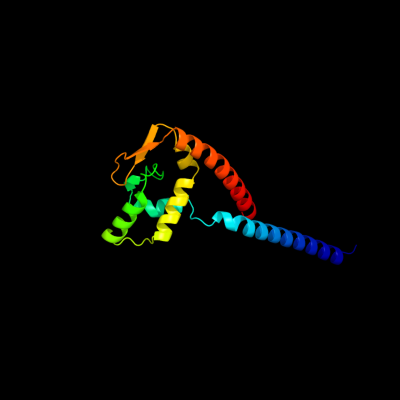
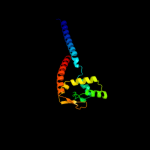
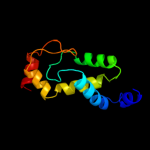
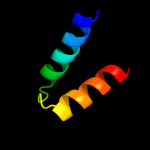
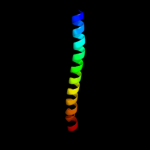
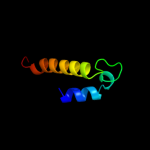
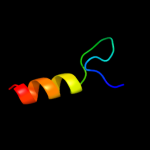
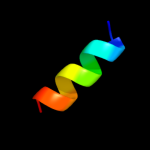
1 d1dvoa_
100.0
97
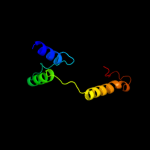
Fold: FinO-likeSuperfamily: FinO-likeFamily: FinO-like2 d2hxja1
100.0
23
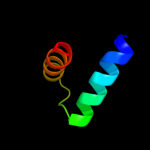
Fold: FinO-likeSuperfamily: FinO-likeFamily: FinO-like3 c2hxjF_
100.0
23
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a protein of unknown function nmb1681 from2 neisseria meningitidis mc58, possible nucleic acid binding protein
4 c2k19A_
53.1
30
PDB header: antimicrobial proteinChain: A: PDB Molecule: putative piscicolin 126 immunity protein;PDBTitle: nmr solution structure of pisi
5 c2zrrA_
37.4
36
PDB header: antimicrobial proteinChain: A: PDB Molecule: mundticin ks immunity protein;PDBTitle: crystal structure of an immunity protein that contributes2 to the self-protection of bacteriocin-producing3 enterococcus mundtii 15-1a
6 d2jdqd1
26.1
37
Fold: PB2 C-terminal domain-likeSuperfamily: PB2 C-terminal domain-likeFamily: PB2 C-terminal domain-like7 c2yicC_
21.3
17
PDB header: lyaseChain: C: PDB Molecule: 2-oxoglutarate decarboxylase;PDBTitle: crystal structure of the suca domain of mycobacterium smegmatis2 alpha-ketoglutarate decarboxylase (triclinic form)
8 d2i5ua1
14.1
15
Fold: DnaD domain-likeSuperfamily: DnaD domain-likeFamily: DnaD domain9 c2w50B_
13.4
9
PDB header: hormoneChain: B: PDB Molecule: armet-like protein 1;PDBTitle: n-terminal domain of human conserved dopamine neurotrophic2 factor (cdnf)
10 c2xt6B_
12.4
17
PDB header: lyaseChain: B: PDB Molecule: 2-oxoglutarate decarboxylase;PDBTitle: crystal structure of mycobacterium smegmatis alpha-ketoglutarate2 decarboxylase homodimer (orthorhombic form)
11 c2w51A_
12.4
14
PDB header: hormoneChain: A: PDB Molecule: protein armet;PDBTitle: human mesencephalic astrocyte-derived neurotrophic factor (2 manf)
12 c3pp5A_
12.3
12
PDB header: structural proteinChain: A: PDB Molecule: brk1;PDBTitle: high-resolution structure of the trimeric scar/wave complex precursor2 brk1
13 d1b33a_
12.1
17
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins14 c3mixA_
12.1
18
PDB header: protein transportChain: A: PDB Molecule: flagellar biosynthesis protein flha;PDBTitle: crystal structure of the cytosolic domain of b. subtilis flha
15 c3a5iB_
11.2
12
PDB header: protein transportChain: B: PDB Molecule: flagellar biosynthesis protein flha;PDBTitle: structure of the cytoplasmic domain of flha
16 c2kq6A_
11.1
25
PDB header: transport proteinChain: A: PDB Molecule: polycystin-2;PDBTitle: the structure of the ef-hand domain of polycystin-2 suggests a2 mechanism for ca2+-dependent regulation of polycystin-2 channel3 activity
17 d1alla_
11.0
17
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins18 c1us2A_
10.9
36
PDB header: hydrolaseChain: A: PDB Molecule: endo-beta-1,4-xylanase;PDBTitle: xylanase10c (mutant e385a) from cellvibrio japonicus in2 complex with xylopentaose
19 d1qnta2
10.7
15
Fold: Ribonuclease H-like motifSuperfamily: Methylated DNA-protein cysteine methyltransferase domainFamily: Methylated DNA-protein cysteine methyltransferase domain20 d1v4va_
10.7
12
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDP-N-acetylglucosamine 2-epimerase21 c3dv9A_
not modelled
9.7
19
PDB header: isomeraseChain: A: PDB Molecule: beta-phosphoglucomutase;PDBTitle: putative beta-phosphoglucomutase from bacteroides vulgatus.
22 d1qh8b_
not modelled
9.6
15
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein23 d1kn1a_
not modelled
9.4
15
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins24 d1cpcb_
not modelled
9.1
23
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins25 c3aicC_
not modelled
8.9
24
PDB header: transferaseChain: C: PDB Molecule: glucosyltransferase-si;PDBTitle: crystal structure of glucansucrase from streptococcus mutans
26 d1gjja2
not modelled
8.9
29
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain27 d1xg0c_
not modelled
8.8
21
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins28 c3mydA_
not modelled
8.4
15
PDB header: protein transportChain: A: PDB Molecule: flagellar biosynthesis protein flha;PDBTitle: structure of the cytoplasmic domain of flha from helicobacter pylori
29 d1ka8a_
not modelled
7.9
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: P4 origin-binding domain-like30 c3hz3A_
not modelled
7.6
35
PDB header: transferaseChain: A: PDB Molecule: glucansucrase;PDBTitle: lactobacillus reuteri n-terminally truncated glucansucrase2 gtf180(d1025n)-sucrose complex
31 c4ktqA_
not modelled
7.2
25
PDB header: transferase/dnaChain: A: PDB Molecule: protein (large fragment of dna polymerase i);PDBTitle: binary complex of the large fragment of dna polymerase i2 from t. aquaticus bound to a primer/template dna
32 d1s9aa_
not modelled
6.8
3
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase33 c3hj8A_
not modelled
6.8
10
PDB header: oxidoreductaseChain: A: PDB Molecule: catechol 1,2-dioxygenase;PDBTitle: crystal structure determination of catechol 1,2-dioxygenase from2 rhodococcus opacus 1cp in complex with 4-chlorocatechol
34 c3s2xB_
not modelled
6.7
12
PDB header: transferaseChain: B: PDB Molecule: acetyl-coa synthase subunit alpha;PDBTitle: structure of acetyl-coenzyme a synthase alpha subunit c-terminal2 domain
35 c1tmxA_
not modelled
6.5
13
PDB header: oxidoreductaseChain: A: PDB Molecule: hydroxyquinol 1,2-dioxygenase;PDBTitle: crystal structure of hydroxyquinol 1,2-dioxygenase from2 nocardioides simplex 3e
36 c2boyC_
not modelled
6.4
7
PDB header: oxidoreductaseChain: C: PDB Molecule: 3-chlorocatechol 1,2-dioxygenase;PDBTitle: crystal structure of 3-chlorocatechol 1,2-dioxygenase from2 rhodococcus opacus 1cp
37 c2c7jB_
not modelled
6.4
21
PDB header: electron transportChain: B: PDB Molecule: phycoerythrocyanin beta chain;PDBTitle: phycoerythrocyanin from mastigocladus laminosus, 295 k,2 3.0 a
38 d1dmha_
not modelled
6.4
10
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase39 c2azqA_
not modelled
6.1
20
PDB header: oxidoreductaseChain: A: PDB Molecule: catechol 1,2-dioxygenase;PDBTitle: crystal structure of catechol 1,2-dioxygenase from pseudomonas arvilla2 c-1
40 d2c7fa2
not modelled
6.0
25
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases41 d2b1xa1
not modelled
6.0
17
Fold: ISP domainSuperfamily: ISP domainFamily: Ring hydroxylating alpha subunit ISP domain42 d2ijqa1
not modelled
5.9
11
Fold: Hyaluronidase domain-likeSuperfamily: TTHA0068-likeFamily: TTHA0068-like43 c2kpqA_
not modelled
5.9
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of agrobacterium tumefaciens protein atu1219:2 northeast structural genomics consortium target att14
44 c2zc2A_
not modelled
5.5
22
PDB header: replicationChain: A: PDB Molecule: dnad-like replication protein;PDBTitle: crystal structure of dnad-like replication protein from2 streptococcus mutans ua159, gi 24377835, residues 127-199
45 c2vmlA_
not modelled
5.5
17
PDB header: photosynthesisChain: A: PDB Molecule: phycocyanin alpha chain;PDBTitle: the monoclinic structure of phycocyanin from gloeobacter2 violaceus
46 d1ezfa_
not modelled
5.5
26
Fold: Terpenoid synthasesSuperfamily: Terpenoid synthasesFamily: Squalene synthase47 d1liab_
not modelled
5.4
23
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins48 d2ghra1
not modelled
5.2
33
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: HTS-like49 d1b8da_
not modelled
5.2
15
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins