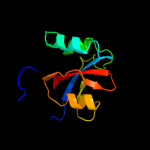
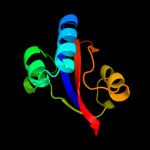
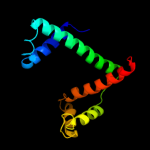
1 c1m5yB_
100.0
22
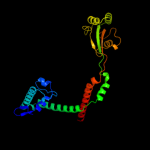
PDB header: isomerase, cell cycleChain: B: PDB Molecule: survival protein sura;PDBTitle: crystallographic structure of sura, a molecular chaperone2 that facilitates outer membrane porin folding
2 c3nrkA_
100.0
21
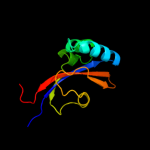
PDB header: unknown functionChain: A: PDB Molecule: lic12922;PDBTitle: the crystal structure of the leptospiral hypothetical protein lic12922
3 c2pv3B_
100.0
26
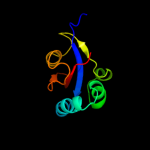
PDB header: isomeraseChain: B: PDB Molecule: chaperone sura;PDBTitle: crystallographic structure of sura fragment lacking the second2 peptidyl-prolyl isomerase domain complexed with peptide nftlkfwdifrk
4 c3rgcB_
100.0
16
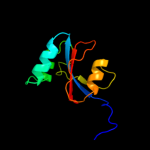
PDB header: chaperoneChain: B: PDB Molecule: possible periplasmic protein;PDBTitle: the virulence factor peb4 and the periplasmic protein cj1289 are two2 structurally related sura-like chaperones in the human pathogen3 campylobacter jejuni
5 c3rfwA_
100.0
26
PDB header: chaperoneChain: A: PDB Molecule: cell-binding factor 2;PDBTitle: the virulence factor peb4 and the periplasmic protein cj1289 are two2 structurally-related sura-like chaperones in the human pathogen3 campylobacter jejuni
6 d1m5ya1
99.9
24
Fold: Triger factor/SurA peptide-binding domain-likeSuperfamily: Triger factor/SurA peptide-binding domain-likeFamily: Porin chaperone SurA, peptide-binding domain7 c2kgjA_
99.8
96
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase d;PDBTitle: solution structure of parvulin domain of ppid from e.coli
8 c2rqsA_
99.8
28
PDB header: isomeraseChain: A: PDB Molecule: parvulin-like peptidyl-prolyl isomerase;PDBTitle: 3d structure of pin from the psychrophilic archeon cenarcheaum2 symbiosum (cspin)
9 d1pina2
99.8
25
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase10 d1jnsa_
99.8
22
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase11 c1yw5A_
99.8
18
PDB header: isomeraseChain: A: PDB Molecule: peptidyl prolyl cis/trans isomerase;PDBTitle: peptidyl-prolyl isomerase ess1 from candida albicans
12 c2jzvA_
99.8
33
PDB header: isomeraseChain: A: PDB Molecule: foldase protein prsa;PDBTitle: solution structure of s. aureus prsa-ppiase
13 d1eq3a_
99.8
20
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase14 d1m5ya3
99.8
31
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase15 c3gpkA_
99.8
32
PDB header: isomeraseChain: A: PDB Molecule: ppic-type peptidyl-prolyl cis-trans isomerase;PDBTitle: crystal structure of ppic-type peptidyl-prolyl cis-trans isomerase2 domain at 1.55a resolution.
16 d1j6ya_
99.8
24
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase17 c1zk6A_
99.8
25
PDB header: isomeraseChain: A: PDB Molecule: foldase protein prsa;PDBTitle: nmr solution structure of b. subtilis prsa ppiase
18 c1f8aB_
99.8
24
PDB header: isomeraseChain: B: PDB Molecule: peptidyl-prolyl cis-trans isomerase nima-PDBTitle: structural basis for the phosphoserine-proline recognition2 by group iv ww domains
19 d2pv2a1
99.7
38
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase20 c3gtyX_
97.2
15
PDB header: chaperone/ribosomal proteinChain: X: PDB Molecule: trigger factor;PDBTitle: promiscuous substrate recognition in folding and assembly activities2 of the trigger factor chaperone
21 c2nsaA_
not modelled
97.1
15
PDB header: chaperoneChain: A: PDB Molecule: trigger factor;PDBTitle: structures of and interactions between domains of trigger factor from2 themotoga maritim
22 c1w26B_
not modelled
96.9
17
PDB header: chaperoneChain: B: PDB Molecule: trigger factor;PDBTitle: trigger factor in complex with the ribosome forms a2 molecular cradle for nascent proteins
23 d1w26a1
not modelled
96.6
17
Fold: Triger factor/SurA peptide-binding domain-likeSuperfamily: Triger factor/SurA peptide-binding domain-likeFamily: TF C-terminus24 c1t11A_
not modelled
94.8
21
PDB header: chaperoneChain: A: PDB Molecule: trigger factor;PDBTitle: trigger factor
25 d1t11a1
not modelled
94.2
21
Fold: Triger factor/SurA peptide-binding domain-likeSuperfamily: Triger factor/SurA peptide-binding domain-likeFamily: TF C-terminus26 c2fcdA_
not modelled
35.0
16
PDB header: cell cycleChain: A: PDB Molecule: myosin light chain 1;PDBTitle: solution structure of n-lobe myosin light chain from2 saccharomices cerevisiae
27 d1wdcb_
not modelled
25.9
14
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like28 c2dguA_
not modelled
20.7
16
PDB header: rna binding proteinChain: A: PDB Molecule: heterogeneous nuclear ribonucleoprotein q;PDBTitle: solution structure of the rna binding domain in2 heterogeneous nuclear ribonucleoprotein q
29 c2osqA_
not modelled
18.0
6
PDB header: rna binding proteinChain: A: PDB Molecule: nucleolar protein 3;PDBTitle: nmr structure of rrm-1 of yeast npl3 protein
30 c3fkkA_
not modelled
15.7
21
PDB header: lyaseChain: A: PDB Molecule: l-2-keto-3-deoxyarabonate dehydratase;PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
31 c2phcB_
not modelled
14.9
36
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein ph0987;PDBTitle: crystal structure of conserved uncharacterized protein ph0987 from2 pyrococcus horikoshii
32 c3mmlD_
not modelled
14.4
18
PDB header: hydrolaseChain: D: PDB Molecule: allophanate hydrolase subunit 1;PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
33 c2dgtA_
not modelled
12.6
6
PDB header: rna binding proteinChain: A: PDB Molecule: rna-binding protein 30;PDBTitle: solution structure of the second rna binding domain in rna-2 binding protein 30
34 c2zp2B_
not modelled
12.5
18
PDB header: transferase inhibitorChain: B: PDB Molecule: kinase a inhibitor;PDBTitle: c-terminal domain of kipi from bacillus subtilis
35 d1wrka1
not modelled
12.1
16
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like36 c3ipdB_
not modelled
11.5
21
PDB header: exocytosisChain: B: PDB Molecule: syntaxin-1a;PDBTitle: helical extension of the neuronal snare complex into the2 membrane, spacegroup i 21 21 21
37 c1slmA_
not modelled
11.1
24
PDB header: hydrolaseChain: A: PDB Molecule: stromelysin-1;PDBTitle: crystal structure of fibroblast stromelysin-1: the c-truncated human2 proenzyme
38 d2phcb1
not modelled
11.1
36
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: PH0987 C-terminal domain-like39 d1u14a_
not modelled
10.9
15
Fold: Anticodon-binding domain-likeSuperfamily: ITPase-likeFamily: YjjX-like40 d1j7ga_
not modelled
10.6
6
Fold: DTD-likeSuperfamily: DTD-likeFamily: DTD-like41 d1sknp_
not modelled
10.4
21
Fold: A DNA-binding domain in eukaryotic transcription factorsSuperfamily: A DNA-binding domain in eukaryotic transcription factorsFamily: A DNA-binding domain in eukaryotic transcription factors42 d1ggwa_
not modelled
10.1
16
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like43 d1zata2
not modelled
10.0
18
Fold: L,D-transpeptidase pre-catalytic domain-likeSuperfamily: L,D-transpeptidase pre-catalytic domain-likeFamily: L,D-transpeptidase pre-catalytic domain-like44 c2dnqA_
not modelled
9.7
12
PDB header: rna binding proteinChain: A: PDB Molecule: rna-binding protein 4b;PDBTitle: solution structure of rna binding domain 1 in rna-binding2 protein 30
45 c2dnpA_
not modelled
9.3
16
PDB header: transcriptionChain: A: PDB Molecule: rna-binding protein 14;PDBTitle: solution structure of rna binding domain 2 in rna-binding2 protein 14
46 c2jvoA_
not modelled
8.6
6
PDB header: rna binding proteinChain: A: PDB Molecule: nucleolar protein 3;PDBTitle: segmental isotope labeling of npl3
47 c2ekwC_
not modelled
8.5
15
PDB header: contractile proteinChain: C: PDB Molecule: myosin catalytic light chain lc-1, mantle muscle;PDBTitle: the crystal structure of squid myosin s1 in the presence of2 so4 2-
48 d1qv0a_
not modelled
8.4
11
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like49 d1oqpa_
not modelled
8.3
20
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like50 d2cxaa1
not modelled
8.1
22
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: LFTR-like51 c2cxaA_
not modelled
8.1
22
PDB header: transferaseChain: A: PDB Molecule: leucyl/phenylalanyl-trna-protein transferase;PDBTitle: crystal structure of leucyl/phenylalanyl-trna protein2 transferase from escherichia coli
52 d1wi6a1
not modelled
8.1
6
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD53 c2kz2A_
not modelled
8.0
21
PDB header: metal binding proteinChain: A: PDB Molecule: calmodulin;PDBTitle: calmodulin, c-terminal domain, f92e mutant
54 c2oy6B_
not modelled
7.8
19
PDB header: contractile proteinChain: B: PDB Molecule: myosin regulatory light chain lc-2;PDBTitle: crystal structure of squid mg.adp myosin s1
55 c2d58A_
not modelled
7.7
12
PDB header: metal binding proteinChain: A: PDB Molecule: allograft inflammatory factor 1;PDBTitle: human microglia-specific protein iba1
56 d2e74h1
not modelled
7.7
11
Fold: Single transmembrane helixSuperfamily: PetN subunit of the cytochrome b6f complexFamily: PetN subunit of the cytochrome b6f complex57 c1vf5H_
not modelled
7.5
13
PDB header: photosynthesisChain: H: PDB Molecule: protein pet n;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
58 c1vf5U_
not modelled
7.5
13
PDB header: photosynthesisChain: U: PDB Molecule: protein pet n;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
59 c2d2cU_
not modelled
7.5
13
PDB header: photosynthesisChain: U: PDB Molecule: cytochrome b6-f complex subunit viii;PDBTitle: crystal structure of cytochrome b6f complex with dbmib from2 m. laminosus
60 c2d2cH_
not modelled
7.5
13
PDB header: photosynthesisChain: H: PDB Molecule: cytochrome b6-f complex subunit viii;PDBTitle: crystal structure of cytochrome b6f complex with dbmib from2 m. laminosus
61 d1fw4a_
not modelled
7.5
20
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like62 c2a4jA_
not modelled
7.5
22
PDB header: structural proteinChain: A: PDB Molecule: centrin 2;PDBTitle: solution structure of the c-terminal domain (t94-y172) of2 the human centrin 2 in complex with a 17 residues peptide3 (p1-xpc) from xeroderma pigmentosum group c protein
63 c3eb2A_
not modelled
7.5
12
PDB header: lyaseChain: A: PDB Molecule: putative dihydrodipicolinate synthetase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
64 c3sibA_
not modelled
7.5
16
PDB header: dna binding proteinChain: A: PDB Molecule: ure3-bp sequence specific dna binding protein;PDBTitle: crystal structure of ure3-binding protein, wild-type
65 c2e76H_
not modelled
7.3
11
PDB header: photosynthesisChain: H: PDB Molecule: cytochrome b6-f complex subunit 8;PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
66 c2e75H_
not modelled
7.3
11
PDB header: photosynthesisChain: H: PDB Molecule: cytochrome b6-f complex subunit 8;PDBTitle: crystal structure of the cytochrome b6f complex with 2-nonyl-4-2 hydroxyquinoline n-oxide (nqno) from m.laminosus
67 c2e74H_
not modelled
7.3
11
PDB header: photosynthesisChain: H: PDB Molecule: cytochrome b6-f complex subunit 8;PDBTitle: crystal structure of the cytochrome b6f complex from m.laminosus
68 d1c07a_
not modelled
7.2
13
Fold: EF Hand-likeSuperfamily: EF-handFamily: Eps15 homology domain (EH domain)69 d2hg7a1
not modelled
7.0
16
Fold: gpW/XkdW-likeSuperfamily: XkdW-likeFamily: XkdW-like70 c2hg7A_
not modelled
7.0
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phage-like element pbsx protein xkdw;PDBTitle: solution nmr structure of phage-like element pbsx protein2 xkdw, northeast structural genomics consortium target sr355
71 c3oepA_
not modelled
6.9
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative uncharacterized protein ttha0988;PDBTitle: crystal structure of ttha0988 in space group p43212
72 d1lkja_
not modelled
6.9
13
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like73 d1wg1a_
not modelled
6.9
6
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD74 d1ap4a_
not modelled
6.8
16
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like75 c2kz5A_
not modelled
6.8
15
PDB header: transcriptionChain: A: PDB Molecule: transcription factor nf-e2 45 kda subunit;PDBTitle: solution nmr structure of transcription factor nf-e2 subunit's dna2 binding domain from homo sapiens, northeast structural genomics3 consortium target hr4653b
76 c1djyB_
not modelled
6.7
10
PDB header: lipid degradationChain: B: PDB Molecule: phosphoinositide-specific phospholipase c,PDBTitle: phosphoinositide-specific phospholipase c-delta1 from rat2 complexed with inositol-2,4,5-trisphosphate
77 c2kgrA_
not modelled
6.7
13
PDB header: protein bindingChain: A: PDB Molecule: intersectin-1;PDBTitle: solution structure of protein itsn1 from homo sapiens.2 northeast structural genomics consortium target hr5524a
78 c1jc2A_
not modelled
6.7
14
PDB header: structural proteinChain: A: PDB Molecule: troponin c, skeletal muscle;PDBTitle: complex of the c-domain of troponin c with residues 1-40 of2 troponin i
79 d1jc2a_
not modelled
6.7
14
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like80 c2amiA_
not modelled
6.6
19
PDB header: cell cycleChain: A: PDB Molecule: caltractin;PDBTitle: solution structure of the calcium-loaded n-terminal sensor2 domain of centrin
81 c1wsuA_
not modelled
6.6
11
PDB header: translation/rnaChain: A: PDB Molecule: selenocysteine-specific elongation factor;PDBTitle: c-terminal domain of elongation factor selb complexed with2 secis rna
82 c3aqpB_
not modelled
6.5
13
PDB header: membrane proteinChain: B: PDB Molecule: probable secdf protein-export membrane protein;PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
83 d1whya_
not modelled
6.5
16
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD84 d1wdcc_
not modelled
6.4
19
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like85 d1whza_
not modelled
6.4
21
Fold: dsRBD-likeSuperfamily: YcfA/nrd intein domainFamily: YcfA-like86 d1stza1
not modelled
6.3
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Heat-inducible transcription repressor HrcA, N-terminal domain87 c2voyB_
not modelled
6.3
18
PDB header: hydrolaseChain: B: PDB Molecule: sarcoplasmic/endoplasmic reticulum calciumPDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
88 c2k7bA_
not modelled
6.2
9
PDB header: metal binding proteinChain: A: PDB Molecule: calcium-binding protein 1;PDBTitle: nmr structure of mg2+-bound cabp1 n-domain
89 c2xnqA_
not modelled
6.1
13
PDB header: rna binding proteinChain: A: PDB Molecule: nuclear polyadenylated rna-binding protein 3;PDBTitle: structural insights into cis element recognition of non-2 polyadenylated rnas by the nab3-rrm
90 c2k29A_
not modelled
6.1
17
PDB header: transcriptionChain: A: PDB Molecule: antitoxin relb;PDBTitle: structure of the dbd domain of e. coli antitoxin relb
91 d1kf6c_
not modelled
6.1
11
Fold: Heme-binding four-helical bundleSuperfamily: Fumarate reductase respiratory complex transmembrane subunitsFamily: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)92 c2ytcA_
not modelled
6.0
17
PDB header: transcriptionChain: A: PDB Molecule: pre-mrna-splicing factor rbm22;PDBTitle: solution structure of rna binding domain in pre-mrna-2 splicing factor rbm22
93 d1whxa_
not modelled
6.0
16
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD94 c2k1lB_
not modelled
6.0
24
PDB header: signaling proteinChain: B: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
95 c2k1kB_
not modelled
6.0
24
PDB header: signaling proteinChain: B: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
96 c2k1kA_
not modelled
6.0
24
PDB header: signaling proteinChain: A: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
97 c2k1lA_
not modelled
6.0
24
PDB header: signaling proteinChain: A: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
98 c3hkzY_
not modelled
6.0
10
PDB header: transferaseChain: Y: PDB Molecule: dna-directed rna polymerase subunit 13;PDBTitle: the x-ray crystal structure of rna polymerase from archaea
99 c2ktgA_
not modelled
6.0
17
PDB header: ca-binding proteinChain: A: PDB Molecule: calmodulin, putative;PDBTitle: calmodulin like protein from entamoeba histolytica: solution structure2 and calcium binding properties of a partially folded protein