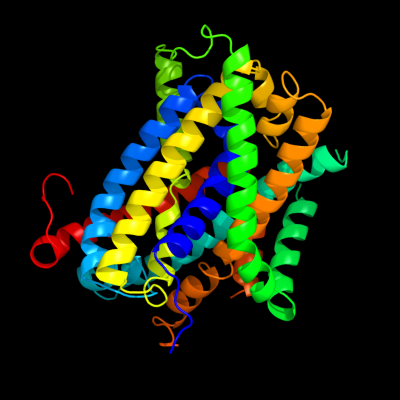
1 c3giaA_
99.9
12
PDB header: transport proteinChain: A: PDB Molecule: uncharacterized protein mj0609;PDBTitle: crystal structure of apct transporter
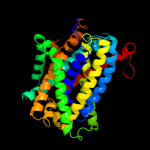
2 c2jlnA_
99.8
8
PDB header: membrane proteinChain: A: PDB Molecule: mhp1;PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
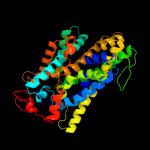
3 c3lrcC_
99.6
15
PDB header: transport proteinChain: C: PDB Molecule: arginine/agmatine antiporter;PDBTitle: structure of e. coli adic (p1)
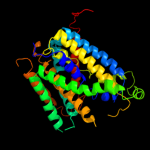
4 c3dh4A_
99.0
10
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
5 c2xq2A_
99.0
11
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: structure of the k294a mutant of vsglt
6 d2a65a1
98.7
15
Fold: SNF-likeSuperfamily: SNF-likeFamily: SNF-like7 c2w8aC_
96.0
14
PDB header: membrane proteinChain: C: PDB Molecule: glycine betaine transporter betp;PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
8 c3hfxA_
94.9
11
PDB header: transport proteinChain: A: PDB Molecule: l-carnitine/gamma-butyrobetaine antiporter;PDBTitle: crystal structure of carnitine transporter
9 d1c8za_
48.2
23
Fold: Tubby C-terminal domain-likeSuperfamily: Tubby C-terminal domain-likeFamily: Transcriptional factor tubby, C-terminal domain10 c2h2wA_
30.8
14
PDB header: transferaseChain: A: PDB Molecule: homoserine o-succinyltransferase;PDBTitle: crystal structure of homoserine o-succinyltransferase (ec 2.3.1.46)2 (homoserine o-transsuccinylase) (hts) (tm0881) from thermotoga3 maritima at 2.52 a resolution
11 d2ghra1
28.3
17
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: HTS-like12 d1ml8a_
26.9
0
Fold: OsmC-likeSuperfamily: OsmC-likeFamily: YhfA-like13 d1vlaa_
22.7
14
Fold: OsmC-likeSuperfamily: OsmC-likeFamily: YhfA-like14 d1htjf_
22.4
19
Fold: Regulator of G-protein signaling, RGSSuperfamily: Regulator of G-protein signaling, RGSFamily: Regulator of G-protein signaling, RGS15 c1htjF_
22.4
19
PDB header: signaling proteinChain: F: PDB Molecule: kiaa0380;PDBTitle: structure of the rgs-like domain from pdz-rhogef
16 c2kncB_
19.2
13
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
17 d2hxva2
18.4
10
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like18 d1fftb2
17.5
8
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region19 c1lqlE_
16.8
22
PDB header: unknown functionChain: E: PDB Molecule: osmotical inducible protein c like family;PDBTitle: crystal structure of osmc like protein from mycoplasma2 pneumoniae
20 d1lqla_
16.8
22
Fold: OsmC-likeSuperfamily: OsmC-likeFamily: Ohr/OsmC resistance proteins21 c2lkgA_
not modelled
16.8
25
PDB header: signaling proteinChain: A: PDB Molecule: acetylcholine receptor;PDBTitle: wsa major conformation
22 c2k21A_
not modelled
16.6
18
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily ePDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
23 d1z3aa1
not modelled
15.4
23
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like24 d1qwia_
not modelled
15.3
14
Fold: OsmC-likeSuperfamily: OsmC-likeFamily: Ohr/OsmC resistance proteins25 c2egtA_
not modelled
14.6
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein aq_1549;PDBTitle: crystal structure of hypothetical protein (aq1549) from aquifex2 aeolicus
26 d2pn2a1
not modelled
13.5
20
Fold: OsmC-likeSuperfamily: OsmC-likeFamily: Ohr/OsmC resistance proteins27 d2b3ja1
not modelled
12.9
11
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like28 c2be6F_
not modelled
12.3
19
PDB header: membrane proteinChain: F: PDB Molecule: voltage-dependent l-type calcium channel alpha-1c subunit;PDBTitle: 2.0 a crystal structure of the cav1.2 iq domain-ca/cam complex
29 c2bjoA_
not modelled
11.5
23
PDB header: oxidoreductaseChain: A: PDB Molecule: organic hydroperoxide resistance protein ohrb;PDBTitle: crystal structure of the organic hydroperoxide resistance2 protein ohrb of bacillus subtilis
30 c3m4wH_
not modelled
10.6
44
PDB header: signaling protein/signaling proteinChain: H: PDB Molecule: sigma-e factor negative regulatory protein;PDBTitle: structural basis for the negative regulation of bacterial stress2 response by rseb
31 c1shzF_
not modelled
8.7
15
PDB header: signaling proteinChain: F: PDB Molecule: rho guanine nucleotide exchange factor 1;PDBTitle: crystal structure of the p115rhogef rgrgs domain in a2 complex with galpha(13):galpha(i1) chimera
32 c3eerA_
not modelled
8.4
14
PDB header: oxidoreductaseChain: A: PDB Molecule: organic hydroperoxide resistance protein, putative;PDBTitle: high resolution structure of putative organic hydroperoxide resistance2 protein from vibrio cholerae o1 biovar eltor str. n16961
33 d2b3za2
not modelled
8.2
15
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like34 d2onfa1
not modelled
8.1
17
Fold: OsmC-likeSuperfamily: OsmC-likeFamily: Ohr/OsmC resistance proteins35 c2kncA_
not modelled
7.9
12
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
36 d2opla1
not modelled
7.7
13
Fold: OsmC-likeSuperfamily: OsmC-likeFamily: Ohr/OsmC resistance proteins37 c2latA_
not modelled
7.4
19
PDB header: membrane proteinChain: A: PDB Molecule: dolichyl-diphosphooligosaccharide--proteinPDBTitle: solution structure of a human minimembrane protein ost4
38 d2d7va1
not modelled
7.4
4
Fold: OsmC-likeSuperfamily: OsmC-likeFamily: Ohr/OsmC resistance proteins39 c2pjwH_
not modelled
7.0
24
PDB header: endocytosis/exocytosisChain: H: PDB Molecule: uncharacterized protein yhl002w;PDBTitle: the vps27/hse1 complex is a gat domain-based scaffold for2 ubiquitin-dependent sorting
40 d2fika2
not modelled
6.9
20
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain41 c1k8vA_
not modelled
6.9
45
PDB header: unknown functionChain: A: PDB Molecule: neuropeptide f;PDBTitle: the nmr-derived conformation of neuropeptide f from2 moniezia expansa
42 d2o34a1
not modelled
6.8
16
Fold: T-foldSuperfamily: ApbE-likeFamily: DVU1097-like43 c1iflA_
not modelled
6.6
16
PDB header: virusChain: A: PDB Molecule: inovirus;PDBTitle: molecular models and structural comparisons of native and2 mutant class i filamentous bacteriophages ff (fd, f1, m13),3 if1 and ike
44 c2ql8A_
not modelled
6.6
22
PDB header: oxidoreductaseChain: A: PDB Molecule: putative redox protein;PDBTitle: crystal structure of a putative redox protein (lsei_0423) from2 lactobacillus casei atcc 334 at 1.50 a resolution
45 c2qy6A_
not modelled
6.4
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0209 protein yfck;PDBTitle: crystal structure of the n-terminal domain of upf0209 protein yfck2 from escherichia coli o157:h7
46 c2kluA_
not modelled
6.3
30
PDB header: immune system, membrane proteinChain: A: PDB Molecule: t-cell surface glycoprotein cd4;PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
47 c3mk7F_
not modelled
6.1
21
PDB header: oxidoreductaseChain: F: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit p;PDBTitle: the structure of cbb3 cytochrome oxidase
48 c3cjeA_
not modelled
6.1
9
PDB header: oxidoreductaseChain: A: PDB Molecule: osmc-like protein;PDBTitle: crystal structure of an osmc-like hydroperoxide resistance protein2 (jann_2040) from jannaschia sp. ccs1 at 1.70 a resolution
49 d1p6oa_
not modelled
6.1
11
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like50 d1iapa_
not modelled
6.0
13
Fold: Regulator of G-protein signaling, RGSSuperfamily: Regulator of G-protein signaling, RGSFamily: Regulator of G-protein signaling, RGS51 c3lm3A_
not modelled
5.9
28
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative glycoside hydrolase/deacetylase2 (bdi_3119) from parabacteroides distasonis at 1.44 a resolution
52 d1uspa_
not modelled
5.8
18
Fold: OsmC-likeSuperfamily: OsmC-likeFamily: Ohr/OsmC resistance proteins53 d2a6aa1
not modelled
5.8
18
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: YeaZ-like54 c2nx8A_
not modelled
5.8
26
PDB header: hydrolaseChain: A: PDB Molecule: trna-specific adenosine deaminase;PDBTitle: the crystal structure of the trna-specific adenosine deaminase from2 streptococcus pyogenes
55 d2po6a2
not modelled
5.4
15
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain56 d1onqa2
not modelled
5.3
15
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain57 c2x58B_
not modelled
5.2
42
PDB header: oxidoreductaseChain: B: PDB Molecule: peroxisomal bifunctional enzyme;PDBTitle: the crystal structure of mfe1 liganded with coa