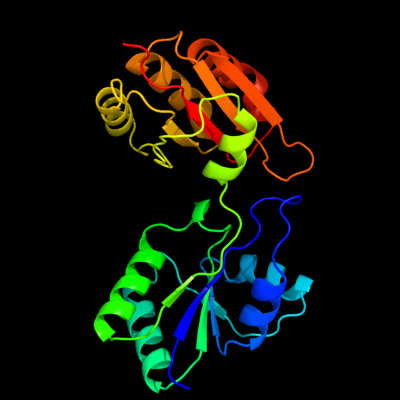
| 1 | c3kwpA_
|
|
 |
100.0 |
49 |
PDB header:transferase
Chain: A: PDB Molecule:predicted methyltransferase;
PDBTitle: crystal structure of putative methyltransferase from lactobacillus2 brevis
|
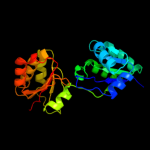
| 2 | d1cbfa_
|
|
 |
100.0 |
17 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
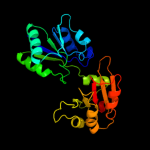
| 3 | c1cbfA_
|
|
 |
100.0 |
17 |
PDB header:methyltransferase
Chain: A: PDB Molecule:cobalt-precorrin-4 transmethylase;
PDBTitle: the x-ray structure of a cobalamin biosynthetic enzyme, cobalt2 precorrin-4 methyltransferase, cbif
|
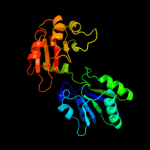
| 4 | c1pjtB_
|
|
 |
100.0 |
20 |
PDB header:transferase/oxidoreductase/lyase
Chain: B: PDB Molecule:siroheme synthase;
PDBTitle: the structure of the ser128ala point-mutant variant of cysg,2 the multifunctional3 methyltransferase/dehydrogenase/ferrochelatase for4 siroheme synthesis
|
| 5 | c2yboA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: the x-ray structure of the sam-dependent uroporphyrinogen2 iii methyltransferase nire from pseudomonas aeruginosa in3 complex with sah
|
| 6 | d1wyza1
|
|
 |
100.0 |
22 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 7 | d1s4da_
|
|
 |
100.0 |
15 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 8 | d1pjqa2
|
|
 |
100.0 |
20 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 9 | c3ndcB_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:precorrin-4 c(11)-methyltransferase;
PDBTitle: crystal structure of precorrin-4 c11-methyltransferase from2 rhodobacter capsulatus
|
| 10 | c2zvbA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-3 c17-methyltransferase;
PDBTitle: crystal structure of tt0207 from thermus thermophilus hb8
|
| 11 | d1va0a1
|
|
 |
100.0 |
14 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 12 | d1wdea_
|
|
 |
100.0 |
18 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 13 | d1ve2a1
|
|
 |
100.0 |
21 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 14 | c3nutC_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: C: PDB Molecule:precorrin-3 methylase;
PDBTitle: crystal structure of the methyltransferase cobj
|
| 15 | c3nd1B_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:precorrin-6a synthase/cobf protein;
PDBTitle: crystal structure of precorrin-6a synthase from rhodobacter capsulatus
|
| 16 | c2qbuA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-2 methyltransferase;
PDBTitle: crystal structure of methanothermobacter thermautotrophicus cbil
|
| 17 | c2e0kA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-2 c20-methyltransferase;
PDBTitle: crystal structure of cbil, a methyltransferase involved in anaerobic2 vitamin b12 biosynthesis
|
| 18 | d1vhva_
|
|
 |
100.0 |
13 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 19 | d2deka1
|
|
 |
100.0 |
21 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 20 | c3i4tA_
|
|
 |
100.0 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:diphthine synthase;
PDBTitle: crystal structure of putative diphthine synthase from2 entamoeba histolytica
|
| 21 | c2npnA_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:putative cobalamin synthesis related protein;
PDBTitle: crystal structure of putative cobalamin synthesis related protein2 (cobf) from corynebacterium diphtheriae
|
| 22 | c2bb3B_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:cobalamin biosynthesis precorrin-6y methylase (cbie);
PDBTitle: crystal structure of cobalamin biosynthesis precorrin-6y methylase2 (cbie) from archaeoglobus fulgidus
|
| 23 | d2bb3a1 |
|
not modelled |
100.0 |
18 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
| 24 | c3fq6A_ |
|
not modelled |
100.0 |
40 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: the crystal structure of a methyltransferase domain from bacteroides2 thetaiotaomicron vpi
|
| 25 | c3hh1D_ |
|
not modelled |
100.0 |
45 |
PDB header:transferase
Chain: D: PDB Molecule:tetrapyrrole methylase family protein;
PDBTitle: the structure of a tetrapyrrole methylase family protein domain from2 chlorobium tepidum tls
|
| 26 | d2ezla_
|
|
 |
85.4 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Recombinase DNA-binding domain |
| 27 | c2px0D_ |
|
not modelled |
75.1 |
15 |
PDB header:biosynthetic protein
Chain: D: PDB Molecule:flagellar biosynthesis protein flhf;
PDBTitle: crystal structure of flhf complexed with gmppnp/mg(2+)
|
| 28 | c3q3vA_ |
|
not modelled |
73.8 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoglycerate kinase;
PDBTitle: crystal structure of phosphoglycerate kinase from campylobacter2 jejuni.
|
| 29 | c1w78A_ |
|
not modelled |
56.4 |
14 |
PDB header:synthase
Chain: A: PDB Molecule:folc bifunctional protein;
PDBTitle: e.coli folc in complex with dhpp and adp
|
| 30 | c2z1dA_ |
|
not modelled |
51.7 |
12 |
PDB header:metal binding protein
Chain: A: PDB Molecule:hydrogenase expression/formation protein hypd;
PDBTitle: crystal structure of [nife] hydrogenase maturation protein, hypd from2 thermococcus kodakaraensis
|
| 31 | d1ii7a_ |
|
not modelled |
51.2 |
20 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DNA double-strand break repair nuclease |
| 32 | c3navB_ |
|
not modelled |
44.0 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
| 33 | c3bioB_ |
|
not modelled |
42.3 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of oxidoreductase (gfo/idh/moca family member) from2 porphyromonas gingivalis w83
|
| 34 | c3l7oB_ |
|
not modelled |
40.2 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from streptococcus2 mutans ua159
|
| 35 | d1ptma_ |
|
not modelled |
39.4 |
19 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
| 36 | c2q5cA_ |
|
not modelled |
38.5 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
| 37 | c2q7xA_ |
|
not modelled |
36.3 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:upf0052 protein sp_1565;
PDBTitle: crystal structure of a putative phospho transferase (sp_1565) from2 streptococcus pneumoniae tigr4 at 2.00 a resolution
|
| 38 | c3av0A_ |
|
not modelled |
35.3 |
16 |
PDB header:recombination
Chain: A: PDB Molecule:dna double-strand break repair protein mre11;
PDBTitle: crystal structure of mre11-rad50 bound to atp s
|
| 39 | d1t9ba1 |
|
not modelled |
34.4 |
14 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 40 | d2hzba1 |
|
not modelled |
34.4 |
10 |
Fold:CofD-like
Superfamily:CofD-like
Family:CofD-like |
| 41 | d1ybha1 |
|
not modelled |
33.4 |
19 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 42 | d1sc6a2 |
|
not modelled |
33.4 |
17 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 43 | d2d59a1 |
|
not modelled |
33.4 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 44 | d1xfia_ |
|
not modelled |
32.9 |
18 |
Fold:AF1104-like
Superfamily:AF1104-like
Family:AF1104-like |
| 45 | c2l0kA_ |
|
not modelled |
31.8 |
26 |
PDB header:transcription
Chain: A: PDB Molecule:stage iii sporulation protein d;
PDBTitle: nmr solution structure of a transcription factor spoiiid in complex2 with dna
|
| 46 | c2z2uA_ |
|
not modelled |
31.7 |
18 |
PDB header:metal binding protein
Chain: A: PDB Molecule:upf0026 protein mj0257;
PDBTitle: crystal structure of archaeal tyw1
|
| 47 | d1gdha2 |
|
not modelled |
30.2 |
17 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 48 | d2naca2 |
|
not modelled |
30.0 |
9 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 49 | c3mvnA_ |
|
not modelled |
29.7 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate:l-alanyl-gamma-d-glutamayl-medo-
PDBTitle: crystal structure of a domain from a putative udp-n-acetylmuramate:l-2 alanyl-gamma-d-glutamayl-medo-diaminopimelate ligase from haemophilus3 ducreyi 35000hp
|
| 50 | c3rhtB_ |
|
not modelled |
29.3 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:(gatase1)-like protein;
PDBTitle: crystal structure of type 1 glutamine amidotransferase (gatase1)-like2 protein from planctomyces limnophilus
|
| 51 | d2z67a1 |
|
not modelled |
29.0 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:SepSecS-like |
| 52 | d1v6ta_ |
|
not modelled |
27.4 |
17 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:LamB/YcsF-like |
| 53 | d2o8ra3 |
|
not modelled |
27.4 |
20 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 54 | c3auzA_ |
|
not modelled |
26.7 |
21 |
PDB header:recombination
Chain: A: PDB Molecule:dna double-strand break repair protein mre11;
PDBTitle: crystal structure of mre11 with manganese
|
| 55 | d2gk3a1 |
|
not modelled |
26.5 |
13 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:STM3548-like |
| 56 | d1xtoa_ |
|
not modelled |
26.4 |
19 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Coenzyme PQQ synthesis protein B, PqqB |
| 57 | c1gqqA_ |
|
not modelled |
25.0 |
18 |
PDB header:cell wall biosynthesis
Chain: A: PDB Molecule:udp-n-acetylmuramate-l-alanine ligase;
PDBTitle: murc - crystal structure of the apo-enzyme from haemophilus2 influenzae
|
| 58 | d1jeyb2 |
|
not modelled |
24.4 |
9 |
Fold:vWA-like
Superfamily:vWA-like
Family:Ku80 subunit N-terminal domain |
| 59 | d1f6ya_ |
|
not modelled |
24.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 60 | d1q6za1 |
|
not modelled |
23.4 |
19 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 61 | c3c19A_ |
|
not modelled |
23.2 |
8 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein mk0293;
PDBTitle: crystal structure of protein mk0293 from methanopyrus kandleri av19
|
| 62 | c3hl2D_ |
|
not modelled |
22.7 |
17 |
PDB header:transferase
Chain: D: PDB Molecule:o-phosphoseryl-trna(sec) selenium transferase;
PDBTitle: the crystal structure of the human sepsecs-trnasec complex
|
| 63 | c2q8uA_ |
|
not modelled |
22.7 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:exonuclease, putative;
PDBTitle: crystal structure of mre11 from thermotoga maritima msb8 (tm1635) at2 2.20 a resolution
|
| 64 | d1ad1a_ |
|
not modelled |
22.6 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 65 | d1ygya2 |
|
not modelled |
22.3 |
17 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 66 | c2nacA_ |
|
not modelled |
22.3 |
10 |
PDB header:oxidoreductase(aldehyde(d),nad+(a))
Chain: A: PDB Molecule:nad-dependent formate dehydrogenase;
PDBTitle: high resolution structures of holo and apo formate dehydrogenase
|
| 67 | c1z4hA_ |
|
not modelled |
22.1 |
13 |
PDB header:protein binding, dna binding protein
Chain: A: PDB Molecule:tor inhibition protein;
PDBTitle: the response regulator tori belongs to a new family of2 atypical excisionase
|
| 68 | c3dmdA_ |
|
not modelled |
21.7 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:signal recognition particle receptor;
PDBTitle: structures and conformations in solution of the signal recognition2 particle receptor from the archaeon pyrococcus furiosus
|
| 69 | c3cf4G_ |
|
not modelled |
21.5 |
13 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:acetyl-coa decarboxylase/synthase epsilon subunit;
PDBTitle: structure of the codh component of the m. barkeri acds complex
|
| 70 | d1qbjc_ |
|
not modelled |
21.2 |
10 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Z-DNA binding domain |
| 71 | d1iuka_ |
|
not modelled |
21.1 |
32 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 72 | c3iwpK_ |
|
not modelled |
20.9 |
13 |
PDB header:metal binding protein
Chain: K: PDB Molecule:copper homeostasis protein cutc homolog;
PDBTitle: crystal structure of human copper homeostasis protein cutc
|
| 73 | d1t57a_ |
|
not modelled |
20.7 |
11 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:MTH1675-like |
| 74 | c3g2bA_ |
|
not modelled |
20.6 |
17 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:coenzyme pqq synthesis protein d;
PDBTitle: crystal structure of pqqd from xanthomonas campestris
|
| 75 | c2yx0A_ |
|
not modelled |
20.2 |
16 |
PDB header:metal binding protein
Chain: A: PDB Molecule:radical sam enzyme;
PDBTitle: crystal structure of p. horikoshii tyw1
|
| 76 | d2jfga2 |
|
not modelled |
20.0 |
21 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 77 | d2djia1 |
|
not modelled |
19.9 |
14 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 78 | c3canA_ |
|
not modelled |
19.4 |
21 |
PDB header:lyase activator
Chain: A: PDB Molecule:pyruvate-formate lyase-activating enzyme;
PDBTitle: crystal structure of a domain of pyruvate-formate lyase-activating2 enzyme from bacteroides vulgatus atcc 8482
|
| 79 | c2qasA_ |
|
not modelled |
19.2 |
20 |
PDB header:hydrolase activator
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of caulobacter crescentus sspb ortholog
|
| 80 | c2ppvA_ |
|
not modelled |
19.1 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a protein belonging to the upf0052 (se_0549) from2 staphylococcus epidermidis atcc 12228 at 2.00 a resolution
|
| 81 | c3qg5D_ |
|
not modelled |
18.5 |
18 |
PDB header:hydrolase
Chain: D: PDB Molecule:mre11;
PDBTitle: the mre11:rad50 complex forms an atp dependent molecular clamp in dna2 double-strand break repair
|
| 82 | d1jeya2 |
|
not modelled |
18.4 |
14 |
Fold:vWA-like
Superfamily:vWA-like
Family:Ku70 subunit N-terminal domain |
| 83 | d1pvda1 |
|
not modelled |
18.3 |
18 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 84 | c3ecsD_ |
|
not modelled |
18.2 |
11 |
PDB header:translation
Chain: D: PDB Molecule:translation initiation factor eif-2b subunit
PDBTitle: crystal structure of human eif2b alpha
|
| 85 | d1p3y1_ |
|
not modelled |
17.9 |
5 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
| 86 | c3lqkA_ |
|
not modelled |
17.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit b;
PDBTitle: crystal structure of dipicolinate synthase subunit b from bacillus2 halodurans c
|
| 87 | c2qazC_ |
|
not modelled |
17.8 |
20 |
PDB header:hydrolase activator
Chain: C: PDB Molecule:sspb protein;
PDBTitle: structure of c. crescentus sspb ortholog
|
| 88 | c3imkA_ |
|
not modelled |
17.5 |
20 |
PDB header:metal binding protein
Chain: A: PDB Molecule:putative molybdenum carrier protein;
PDBTitle: crystal structure of putative molybdenum carrier protein (yp_461806.1)2 from syntrophus aciditrophicus sb at 1.45 a resolution
|
| 89 | c2pjmA_ |
|
not modelled |
16.8 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: structure of ribose 5-phosphate isomerase a from2 methanocaldococcus jannaschii
|
| 90 | c3pfnB_ |
|
not modelled |
16.6 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:nad kinase;
PDBTitle: crystal structure of human nad kinase
|
| 91 | c3oa2B_ |
|
not modelled |
16.3 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:wbpb;
PDBTitle: crystal structure of the wlba (wbpb) dehydrogenase from pseudomonas2 aeruginosa in complex with nad at 1.5 angstrom resolution
|
| 92 | c1yirA_ |
|
not modelled |
16.1 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase 2;
PDBTitle: crystal structure of a nicotinate phosphoribosyltransferase
|
| 93 | d1t6t1_ |
|
not modelled |
16.0 |
10 |
Fold:Toprim domain
Superfamily:Toprim domain
Family:Toprim domain |
| 94 | d1qgpa_ |
|
not modelled |
16.0 |
11 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Z-DNA binding domain |
| 95 | d2ez9a1 |
|
not modelled |
15.6 |
14 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 96 | c1lc3A_ |
|
not modelled |
15.5 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:biliverdin reductase a;
PDBTitle: crystal structure of a biliverdin reductase enzyme-cofactor2 complex
|
| 97 | d2gxba1 |
|
not modelled |
15.1 |
10 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Z-DNA binding domain |
| 98 | d2dlda2 |
|
not modelled |
15.0 |
20 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 99 | d2q4qa1 |
|
not modelled |
15.0 |
16 |
Fold:MTH938-like
Superfamily:MTH938-like
Family:MTH938-like |