| 1 |
|
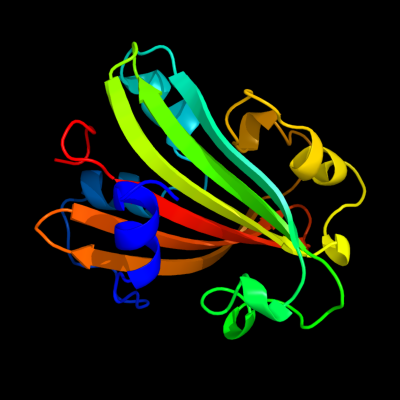
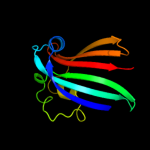
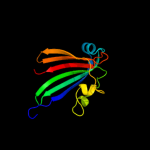
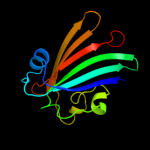
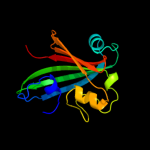
PDB 1tt8 chain A
Region: 2 - 165
Aligned: 164
Modelled: 164
Confidence: 100.0%
Identity: 99%
Fold: Chorismate lyase-like
Superfamily: Chorismate lyase-like
Family: Chorismate lyase
Phyre2
| 2 |
|
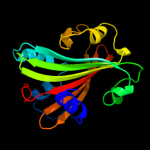
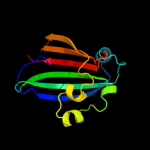
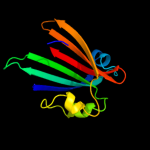
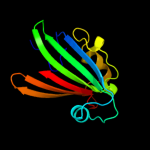
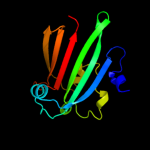
PDB 2nwi chain A domain 1
Region: 21 - 161
Aligned: 141
Modelled: 141
Confidence: 100.0%
Identity: 21%
Fold: Chorismate lyase-like
Superfamily: Chorismate lyase-like
Family: AF1396-like
Phyre2
| 3 |
|
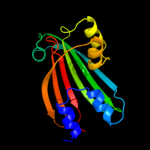
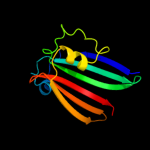
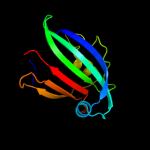
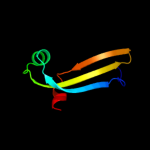
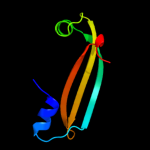
PDB 3f8l chain C
Region: 29 - 161
Aligned: 131
Modelled: 133
Confidence: 98.4%
Identity: 16%
PDB header:transcription
Chain: C: PDB Molecule:hth-type transcriptional repressor phnf;
PDBTitle: crystal structure of the effector domain of phnf from mycobacterium2 smegmatis
Phyre2
| 4 |
|
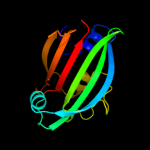
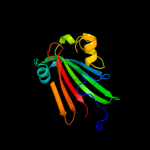
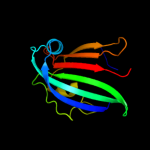
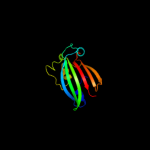
PDB 3hfi chain A
Region: 47 - 161
Aligned: 113
Modelled: 115
Confidence: 97.8%
Identity: 12%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative regulator;
PDBTitle: the crystal structure of the putative regulator from escherichia coli2 cft073
Phyre2
| 5 |
|
PDB 3cnv chain A domain 1
Region: 39 - 161
Aligned: 123
Modelled: 123
Confidence: 97.8%
Identity: 11%
Fold: Chorismate lyase-like
Superfamily: Chorismate lyase-like
Family: UTRA domain
Phyre2
| 6 |
|
PDB 2pkh chain A domain 1
Region: 45 - 161
Aligned: 115
Modelled: 117
Confidence: 97.6%
Identity: 12%
Fold: Chorismate lyase-like
Superfamily: Chorismate lyase-like
Family: UTRA domain
Phyre2
| 7 |
|
PDB 2fa1 chain A domain 1
Region: 29 - 161
Aligned: 130
Modelled: 133
Confidence: 97.6%
Identity: 13%
Fold: Chorismate lyase-like
Superfamily: Chorismate lyase-like
Family: UTRA domain
Phyre2
| 8 |
|
PDB 2p19 chain A domain 1
Region: 40 - 161
Aligned: 119
Modelled: 122
Confidence: 97.4%
Identity: 17%
Fold: Chorismate lyase-like
Superfamily: Chorismate lyase-like
Family: UTRA domain
Phyre2
| 9 |
|
PDB 2ikk chain A domain 1
Region: 45 - 161
Aligned: 115
Modelled: 114
Confidence: 97.4%
Identity: 12%
Fold: Chorismate lyase-like
Superfamily: Chorismate lyase-like
Family: UTRA domain
Phyre2
| 10 |
|
PDB 2ogg chain A domain 1
Region: 49 - 161
Aligned: 110
Modelled: 113
Confidence: 97.2%
Identity: 15%
Fold: Chorismate lyase-like
Superfamily: Chorismate lyase-like
Family: UTRA domain
Phyre2
| 11 |
|
PDB 2ra5 chain A domain 1
Region: 41 - 161
Aligned: 118
Modelled: 121
Confidence: 97.0%
Identity: 18%
Fold: Chorismate lyase-like
Superfamily: Chorismate lyase-like
Family: UTRA domain
Phyre2
| 12 |
|
PDB 2ra5 chain A
Region: 41 - 161
Aligned: 118
Modelled: 121
Confidence: 97.0%
Identity: 18%
PDB header:transcription
Chain: A: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of the putative transcriptional regulator2 from streptomyces coelicolor
Phyre2
| 13 |
|
PDB 3bwg chain A domain 2
Region: 45 - 165
Aligned: 118
Modelled: 121
Confidence: 96.9%
Identity: 8%
Fold: Chorismate lyase-like
Superfamily: Chorismate lyase-like
Family: UTRA domain
Phyre2
| 14 |
|
PDB 2ooi chain A domain 1
Region: 33 - 158
Aligned: 123
Modelled: 126
Confidence: 96.9%
Identity: 11%
Fold: Chorismate lyase-like
Superfamily: Chorismate lyase-like
Family: UTRA domain
Phyre2
| 15 |
|
PDB 3ddv chain A domain 1
Region: 47 - 161
Aligned: 94
Modelled: 94
Confidence: 96.3%
Identity: 19%
Fold: Chorismate lyase-like
Superfamily: Chorismate lyase-like
Family: UTRA domain
Phyre2
| 16 |
|
PDB 3lhe chain A
Region: 42 - 102
Aligned: 61
Modelled: 61
Confidence: 95.1%
Identity: 21%
PDB header:transcription regulator
Chain: A: PDB Molecule:gntr family transcriptional regulator;
PDBTitle: the crystal structure of the c-terminal domain of a gntr2 family transcriptional regulator from bacillus anthracis3 str. sterne
Phyre2
| 17 |
|
PDB 3f8m chain A
Region: 30 - 161
Aligned: 130
Modelled: 132
Confidence: 84.3%
Identity: 16%
PDB header:transcription
Chain: A: PDB Molecule:gntr-family protein transcriptional regulator;
PDBTitle: crystal structure of phnf from mycobacterium smegmatis
Phyre2
| 18 |
|
PDB 3eet chain A
Region: 31 - 161
Aligned: 131
Modelled: 131
Confidence: 80.9%
Identity: 11%
PDB header:transcription regulator
Chain: A: PDB Molecule:putative gntr-family transcriptional regulator;
PDBTitle: crystal structure of putative gntr-family transcriptional2 regulator
Phyre2
| 19 |
|
PDB 3edp chain B
Region: 36 - 161
Aligned: 123
Modelled: 126
Confidence: 57.7%
Identity: 9%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:lin2111 protein;
PDBTitle: the crystal structure of the protein lin2111 (functionally unknown)2 from listeria innocua clip11262
Phyre2
| 20 |
|
PDB 2wv0 chain H
Region: 32 - 101
Aligned: 70
Modelled: 70
Confidence: 35.7%
Identity: 10%
PDB header:transcription
Chain: H: PDB Molecule:hth-type transcriptional repressor yvoa;
PDBTitle: crystal structure of the gntr-hutc family member yvoa from2 bacillus subtilis
Phyre2
| 21 |
|
| 22 |
|