| 1 |
|
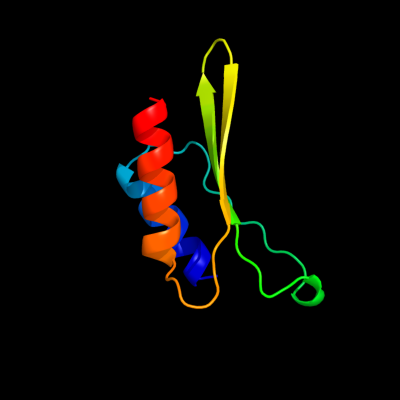
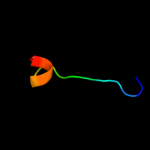
PDB 1qmh chain A domain 1
Region: 37 - 109
Aligned: 73
Modelled: 73
Confidence: 78.0%
Identity: 3%
Fold: Thioredoxin fold
Superfamily: RNA 3'-terminal phosphate cyclase, RPTC, insert domain
Family: RNA 3'-terminal phosphate cyclase, RPTC, insert domain
Phyre2
| 2 |
|
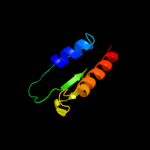
PDB 3fcg chain B
Region: 66 - 90
Aligned: 25
Modelled: 25
Confidence: 54.2%
Identity: 8%
PDB header:membrane protein, protein transport
Chain: B: PDB Molecule:f1 capsule-anchoring protein;
PDBTitle: crystal structure analysis of the middle domain of the2 caf1a usher
Phyre2
| 3 |
|
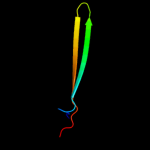
PDB 1ixc chain A domain 1
Region: 19 - 49
Aligned: 31
Modelled: 29
Confidence: 17.3%
Identity: 19%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: LysR-like transcriptional regulators
Phyre2
| 4 |
|
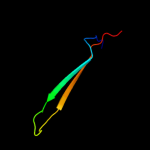
PDB 3ohn chain A
Region: 64 - 90
Aligned: 27
Modelled: 27
Confidence: 17.0%
Identity: 26%
PDB header:membrane protein
Chain: A: PDB Molecule:outer membrane usher protein fimd;
PDBTitle: crystal structure of the fimd translocation domain
Phyre2
| 5 |
|
PDB 2vqi chain A
Region: 64 - 90
Aligned: 27
Modelled: 27
Confidence: 16.7%
Identity: 7%
PDB header:transport
Chain: A: PDB Molecule:outer membrane usher protein papc;
PDBTitle: structure of the p pilus usher (papc) translocation pore
Phyre2
| 6 |
|
PDB 2lf0 chain A
Region: 27 - 42
Aligned: 16
Modelled: 16
Confidence: 15.2%
Identity: 31%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein yibl;
PDBTitle: solution structure of sf3636, a two-domain unknown function protein2 from shigella flexneri 2a, determined by joint refinement of nmr,3 residual dipolar couplings and small-angle x-ray scatting, nesg4 target sfr339/ocsp target sf3636
Phyre2
| 7 |
|
PDB 3rfz chain B
Region: 64 - 90
Aligned: 27
Modelled: 27
Confidence: 13.3%
Identity: 26%
PDB header:cell adhesion/transport/chaperone
Chain: B: PDB Molecule:outer membrane usher protein, type 1 fimbrial synthesis;
PDBTitle: crystal structure of the fimd usher bound to its cognate fimc:fimh2 substrate
Phyre2
| 8 |
|
PDB 2npb chain A
Region: 44 - 113
Aligned: 70
Modelled: 70
Confidence: 12.7%
Identity: 19%
PDB header:oxidoreductase
Chain: A: PDB Molecule:selenoprotein w;
PDBTitle: nmr solution structure of mouse selw
Phyre2
| 9 |
|
PDB 1vyt chain E
Region: 1 - 12
Aligned: 12
Modelled: 12
Confidence: 10.8%
Identity: 58%
PDB header:transport protein
Chain: E: PDB Molecule:voltage-dependent l-type calcium channel
PDBTitle: beta3 subunit complexed with aid
Phyre2
| 10 |
|
PDB 2eny chain A
Region: 58 - 70
Aligned: 13
Modelled: 13
Confidence: 10.3%
Identity: 31%
PDB header:contractile protein
Chain: A: PDB Molecule:obscurin;
PDBTitle: solution structure of the ig-like domain (2735-2825) of2 human obscurin
Phyre2
| 11 |
|
PDB 1bw8 chain A
Region: 51 - 88
Aligned: 38
Modelled: 38
Confidence: 8.5%
Identity: 26%
PDB header:peptide binding protein
Chain: A: PDB Molecule:protein (mu2 adaptin subunit);
PDBTitle: mu2 adaptin subunit (ap50) of ap2 adaptor (second domain),2 complexed with egfr internalization peptide fyralm
Phyre2
| 12 |
|
PDB 2pr9 chain A domain 1
Region: 51 - 88
Aligned: 38
Modelled: 38
Confidence: 7.4%
Identity: 26%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Second domain of Mu2 adaptin subunit (ap50) of ap2 adaptor
Family: Second domain of Mu2 adaptin subunit (ap50) of ap2 adaptor
Phyre2
| 13 |
|
PDB 1vyt chain F
Region: 1 - 14
Aligned: 14
Modelled: 12
Confidence: 7.3%
Identity: 50%
PDB header:transport protein
Chain: F: PDB Molecule:voltage-dependent l-type calcium channel
PDBTitle: beta3 subunit complexed with aid
Phyre2
| 14 |
|
PDB 2wtg chain A
Region: 32 - 51
Aligned: 20
Modelled: 20
Confidence: 6.4%
Identity: 30%
PDB header:oxygen transport
Chain: A: PDB Molecule:globin-like protein;
PDBTitle: high resolution 3d structure of c.elegans globin-like2 protein glb-1
Phyre2
| 15 |
|
PDB 3q13 chain A
Region: 64 - 99
Aligned: 36
Modelled: 36
Confidence: 6.1%
Identity: 8%
PDB header:cell adhesion
Chain: A: PDB Molecule:spondin-1;
PDBTitle: the structure of the ca2+-binding, glycosylated f-spondin domain of f-2 spondin, a c2-domain variant from extracellular matrix
Phyre2
| 16 |
|
PDB 1zh5 chain A domain 1
Region: 11 - 24
Aligned: 14
Modelled: 14
Confidence: 5.8%
Identity: 29%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: La domain
Phyre2
| 17 |
|
PDB 1wu9 chain A domain 1
Region: 41 - 45
Aligned: 5
Modelled: 5
Confidence: 5.8%
Identity: 60%
Fold: EB1 dimerisation domain-like
Superfamily: EB1 dimerisation domain-like
Family: EB1 dimerisation domain-like
Phyre2
| 18 |
|
PDB 3gr1 chain A
Region: 27 - 62
Aligned: 35
Modelled: 36
Confidence: 5.3%
Identity: 17%
PDB header:membrane protein
Chain: A: PDB Molecule:protein prgh;
PDBTitle: periplamic domain of the t3ss inner membrane protein prgh2 from s.typhimurium (fragment 170-392)
Phyre2
| 19 |
|
PDB 2dag chain A
Region: 95 - 128
Aligned: 34
Modelled: 34
Confidence: 5.1%
Identity: 12%
PDB header:hydrolase
Chain: A: PDB Molecule:ubiquitin carboxyl-terminal hydrolase 5;
PDBTitle: solution structure of the first uba domain in the human2 ubiquitin specific protease 5 (isopeptidase 5)
Phyre2
| 20 |
|
PDB 1nw1 chain A
Region: 20 - 108
Aligned: 89
Modelled: 89
Confidence: 5.1%
Identity: 9%
PDB header:transferase
Chain: A: PDB Molecule:choline kinase (49.2 kd);
PDBTitle: crystal structure of choline kinase
Phyre2
| 21 |
|