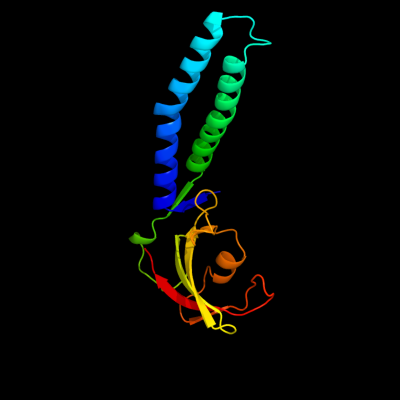
| 1 | c2p4vA_
|
|
 |
100.0 |
35 |
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation factor greb;
PDBTitle: crystal structure of the transcript cleavage factor, greb2 at 2.6a resolution
|
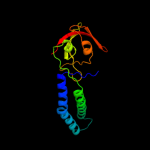
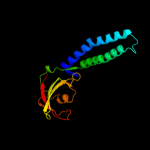
| 2 | c1grjA_
|
|
 |
100.0 |
100 |
PDB header:transcription regulation
Chain: A: PDB Molecule:grea protein;
PDBTitle: grea transcript cleavage factor from escherichia coli
|
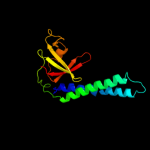
| 3 | c2etnA_
|
|
 |
100.0 |
26 |
PDB header:transcription
Chain: A: PDB Molecule:anti-cleavage anti-grea transcription factor
PDBTitle: crystal structure of thermus aquaticus gfh1
|
| 4 | c3bmbB_
|
|
 |
100.0 |
25 |
PDB header:rna binding protein
Chain: B: PDB Molecule:regulator of nucleoside diphosphate kinase;
PDBTitle: crystal structure of a new rna polymerase interacting2 protein
|
| 5 | c2pn0D_
|
|
 |
100.0 |
29 |
PDB header:transcription
Chain: D: PDB Molecule:prokaryotic transcription elongation factor
PDBTitle: prokaryotic transcription elongation factor grea/greb from2 nitrosomonas europaea
|
| 6 | d1grja1
|
|
 |
99.9 |
100 |
Fold:Long alpha-hairpin
Superfamily:GreA transcript cleavage protein, N-terminal domain
Family:GreA transcript cleavage protein, N-terminal domain |
| 7 | d2f23a1
|
|
 |
99.9 |
26 |
Fold:Long alpha-hairpin
Superfamily:GreA transcript cleavage protein, N-terminal domain
Family:GreA transcript cleavage protein, N-terminal domain |
| 8 | d2f23a2
|
|
 |
99.9 |
28 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:GreA transcript cleavage factor, C-terminal domain |
| 9 | d1grja2
|
|
 |
99.9 |
100 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:GreA transcript cleavage factor, C-terminal domain |
| 10 | d2etna2
|
|
 |
99.9 |
28 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:GreA transcript cleavage factor, C-terminal domain |
| 11 | c3gtyX_
|
|
 |
95.2 |
22 |
PDB header:chaperone/ribosomal protein
Chain: X: PDB Molecule:trigger factor;
PDBTitle: promiscuous substrate recognition in folding and assembly activities2 of the trigger factor chaperone
|
| 12 | d1t11a3
|
|
 |
88.4 |
11 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 13 | d1w26a3
|
|
 |
86.9 |
12 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 14 | c1w26B_
|
|
 |
82.9 |
16 |
PDB header:chaperone
Chain: B: PDB Molecule:trigger factor;
PDBTitle: trigger factor in complex with the ribosome forms a2 molecular cradle for nascent proteins
|
| 15 | c1t11A_
|
|
 |
79.0 |
12 |
PDB header:chaperone
Chain: A: PDB Molecule:trigger factor;
PDBTitle: trigger factor
|
| 16 | d1krha1
|
|
 |
78.6 |
15 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 17 | d1l1pa_
|
|
 |
73.8 |
16 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 18 | d1qx4a1
|
|
 |
63.5 |
13 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 19 | d1ndha1
|
|
 |
59.5 |
8 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 20 | d2cnda1
|
|
 |
58.3 |
17 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 21 | c1zeqX_ |
|
not modelled |
47.4 |
7 |
PDB header:metal binding protein
Chain: X: PDB Molecule:cation efflux system protein cusf;
PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
|
| 22 | c2kw8A_ |
|
not modelled |
45.6 |
18 |
PDB header:protein binding
Chain: A: PDB Molecule:lpxtg-site transpeptidase family protein;
PDBTitle: solution structure of bacillus anthracis sortase a (srta)2 transpeptidase
|
| 23 | d1e32a3 |
|
not modelled |
45.5 |
21 |
Fold:Cdc48 domain 2-like
Superfamily:Cdc48 domain 2-like
Family:Cdc48 domain 2-like |
| 24 | c2gpjA_ |
|
not modelled |
43.9 |
17 |
PDB header:fad-binding protein
Chain: A: PDB Molecule:siderophore-interacting protein;
PDBTitle: crystal structure of a siderophore-interacting protein (sputcn32_0076)2 from shewanella putrefaciens cn-32 at 2.20 a resolution
|
| 25 | c2kfwA_ |
|
not modelled |
41.9 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:fkbp-type peptidyl-prolyl cis-trans isomerase
PDBTitle: solution structure of full-length slyd from e.coli
|
| 26 | d1tvca1 |
|
not modelled |
41.8 |
12 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 27 | d1umka1 |
|
not modelled |
41.6 |
11 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 28 | c2dq0A_ |
|
not modelled |
40.1 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:seryl-trna synthetase;
PDBTitle: crystal structure of seryl-trna synthetase from pyrococcus2 horikoshii complexed with a seryl-adenylate analog
|
| 29 | c3fn5B_ |
|
not modelled |
39.3 |
10 |
PDB header:hydrolase
Chain: B: PDB Molecule:sortase a;
PDBTitle: crystal structure of sortase a (spy1154) from streptococcus2 pyogenes serotype m1 strain sf370
|
| 30 | c3shwA_ |
|
not modelled |
38.5 |
19 |
PDB header:cell adhesion
Chain: A: PDB Molecule:tight junction protein zo-1;
PDBTitle: crystal structure of zo-1 pdz3-sh3-guk supramodule complex with2 connexin-45 peptide
|
| 31 | c3g5oA_ |
|
not modelled |
38.5 |
26 |
PDB header:toxin/antitoxin
Chain: A: PDB Molecule:uncharacterized protein rv2865;
PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
|
| 32 | d1hxva_ |
|
not modelled |
37.3 |
8 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 33 | c1hxvA_ |
|
not modelled |
37.3 |
8 |
PDB header:chaperone
Chain: A: PDB Molecule:trigger factor;
PDBTitle: ppiase domain of the mycoplasma genitalium trigger factor
|
| 34 | d1t2wa_ |
|
not modelled |
36.2 |
11 |
Fold:Sortase
Superfamily:Sortase
Family:Sortase |
| 35 | d1st6a4 |
|
not modelled |
34.3 |
29 |
Fold:Four-helical up-and-down bundle
Superfamily:alpha-catenin/vinculin-like
Family:alpha-catenin/vinculin |
| 36 | c2krgA_ |
|
not modelled |
33.9 |
11 |
PDB header:signaling protein
Chain: A: PDB Molecule:na(+)/h(+) exchange regulatory cofactor nhe-rf1;
PDBTitle: solution structure of human sodium/ hydrogen exchange2 regulatory factor 1(150-358)
|
| 37 | c3shuB_ |
|
not modelled |
33.5 |
26 |
PDB header:cell adhesion
Chain: B: PDB Molecule:tight junction protein zo-1;
PDBTitle: crystal structure of zo-1 pdz3
|
| 38 | d1fdra1 |
|
not modelled |
33.2 |
14 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 39 | d1gvha2 |
|
not modelled |
32.7 |
17 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 40 | c3o0pA_ |
|
not modelled |
32.5 |
14 |
PDB header:transferase , hydrolase
Chain: A: PDB Molecule:sortase family protein;
PDBTitle: pilus-related sortase c of group b streptococcus
|
| 41 | c3re9A_ |
|
not modelled |
31.1 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:sortase-like protein;
PDBTitle: crystal structure of sortasec1 from streptococcus suis
|
| 42 | c2l55A_ |
|
not modelled |
31.0 |
14 |
PDB header:metal binding protein
Chain: A: PDB Molecule:silb,silver efflux protein, mfp component of the three
PDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
|
| 43 | c3cgnA_ |
|
not modelled |
30.8 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:peptidyl-prolyl cis-trans isomerase;
PDBTitle: crystal structure of thermophilic slyd
|
| 44 | c2pjhB_ |
|
not modelled |
30.4 |
13 |
PDB header:transport protein
Chain: B: PDB Molecule:transitional endoplasmic reticulum atpase;
PDBTitle: strctural model of the p97 n domain- npl4 ubd complex
|
| 45 | d1st6a3 |
|
not modelled |
30.0 |
28 |
Fold:Four-helical up-and-down bundle
Superfamily:alpha-catenin/vinculin-like
Family:alpha-catenin/vinculin |
| 46 | c3pr9A_ |
|
not modelled |
29.2 |
8 |
PDB header:chaperone
Chain: A: PDB Molecule:fkbp-type peptidyl-prolyl cis-trans isomerase;
PDBTitle: structural analysis of protein folding by the methanococcus jannaschii2 chaperone fkbp26
|
| 47 | c2kr7A_ |
|
not modelled |
29.1 |
0 |
PDB header:isomerase
Chain: A: PDB Molecule:fkbp-type peptidyl-prolyl cis-trans isomerase slyd;
PDBTitle: solution structure of helicobacter pylori slyd
|
| 48 | c2w1kB_ |
|
not modelled |
28.8 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:putative sortase;
PDBTitle: crystal structure of sortase c-3 (srtc-3) from2 streptococcus pneumoniae
|
| 49 | d1dm9a_ |
|
not modelled |
26.9 |
14 |
Fold:Alpha-L RNA-binding motif
Superfamily:Alpha-L RNA-binding motif
Family:Heat shock protein 15 kD |
| 50 | c1dm9A_ |
|
not modelled |
26.9 |
14 |
PDB header:structural genomics
Chain: A: PDB Molecule:hypothetical 15.5 kd protein in mrca-pcka
PDBTitle: heat shock protein 15 kd
|
| 51 | d2piaa1 |
|
not modelled |
26.9 |
23 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 52 | c2k8iA_ |
|
not modelled |
26.8 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:peptidyl-prolyl cis-trans isomerase;
PDBTitle: solution structure of e.coli slyd
|
| 53 | c2xwgA_ |
|
not modelled |
26.8 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:sortase;
PDBTitle: crystal structure of sortase c-1 from actinomyces oris (formerly2 actinomyces naeslundii)
|
| 54 | d2f5ya1 |
|
not modelled |
26.5 |
19 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 55 | c3k1rA_ |
|
not modelled |
26.0 |
12 |
PDB header:structural protein
Chain: A: PDB Molecule:harmonin;
PDBTitle: structure of harmonin npdz1 in complex with the sam-pbm of2 sans
|
| 56 | c3g66A_ |
|
not modelled |
25.0 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:sortase c;
PDBTitle: the crystal structure of streptococcus pneumoniae sortase c2 provides novel insights into catalysis as well as pilin3 substrate specificity
|
| 57 | d2gp4a1 |
|
not modelled |
24.7 |
11 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LeuD/IlvD-like
Family:IlvD/EDD C-terminal domain-like |
| 58 | d1t9fa_ |
|
not modelled |
23.9 |
10 |
Fold:beta-Trefoil
Superfamily:MIR domain
Family:MIR domain |
| 59 | d1ep3b1 |
|
not modelled |
23.4 |
15 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 60 | d1uita_ |
|
not modelled |
23.4 |
22 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 61 | d1y7na1 |
|
not modelled |
22.8 |
14 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 62 | c3rccI_ |
|
not modelled |
22.7 |
7 |
PDB header:hydrolase
Chain: I: PDB Molecule:sortase srta;
PDBTitle: crystal structure of the streptococcus agalactiae sortase a
|
| 63 | c1cz5A_ |
|
not modelled |
21.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:vcp-like atpase;
PDBTitle: nmr structure of vat-n: the n-terminal domain of vat (vcp-2 like atpase of thermoplasma)
|
| 64 | c3prdA_ |
|
not modelled |
20.8 |
8 |
PDB header:chaperone, isomerase
Chain: A: PDB Molecule:fkbp-type peptidyl-prolyl cis-trans isomerase;
PDBTitle: structural analysis of protein folding by the methanococcus jannaschii2 chaperone fkbp26
|
| 65 | c3u5cG_ |
|
not modelled |
20.3 |
14 |
PDB header:ribosome
Chain: G: PDB Molecule:40s ribosomal protein s6-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution
|
| 66 | d1pina2 |
|
not modelled |
20.3 |
12 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 67 | d1fnda1 |
|
not modelled |
19.3 |
24 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 68 | c2piaA_ |
|
not modelled |
18.1 |
12 |
PDB header:reductase
Chain: A: PDB Molecule:phthalate dioxygenase reductase;
PDBTitle: phthalate dioxygenase reductase: a modular structure for2 electron transfer from pyridine nucleotides to [2fe-2s]
|
| 69 | d2iv2x1 |
|
not modelled |
18.0 |
13 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 70 | c2vn1A_ |
|
not modelled |
17.9 |
10 |
PDB header:isomerase
Chain: A: PDB Molecule:70 kda peptidylprolyl isomerase;
PDBTitle: crystal structure of the fk506-binding domain of plasmodium2 falciparum fkbp35 in complex with fk506
|
| 71 | d1ix5a_ |
|
not modelled |
17.9 |
17 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 72 | d1sm4a1 |
|
not modelled |
17.2 |
25 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 73 | d2a6qa1 |
|
not modelled |
16.9 |
21 |
Fold:YefM-like
Superfamily:YefM-like
Family:YefM-like |
| 74 | c2xznY_ |
|
not modelled |
16.8 |
17 |
PDB header:ribosome
Chain: Y: PDB Molecule:rps6e;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|
| 75 | d2jioa1 |
|
not modelled |
16.7 |
17 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 76 | c1krhA_ |
|
not modelled |
16.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:benzoate 1,2-dioxygenase reductase;
PDBTitle: x-ray stucture of benzoate dioxygenase reductase
|
| 77 | c2gp4A_ |
|
not modelled |
16.0 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:6-phosphogluconate dehydratase;
PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
|
| 78 | c2eehA_ |
|
not modelled |
15.8 |
18 |
PDB header:metal binding protein
Chain: A: PDB Molecule:pdz domain-containing protein 7;
PDBTitle: solution structure of first pdz domain of pdz domain2 containing protein 7
|
| 79 | c2z17A_ |
|
not modelled |
15.7 |
26 |
PDB header:protein binding
Chain: A: PDB Molecule:pleckstrin homology sec7 and coiled-coil domains-
PDBTitle: crystal sturcture of pdz domain from human pleckstrin2 homology, sec7
|
| 80 | d1vloa1 |
|
not modelled |
15.3 |
15 |
Fold:Elongation factor/aminomethyltransferase common domain
Superfamily:Aminomethyltransferase beta-barrel domain
Family:Aminomethyltransferase beta-barrel domain |
| 81 | c3rbjB_ |
|
not modelled |
15.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:sortase family protein;
PDBTitle: crystal structure of the lid-mutant of streptococcus agalactiae2 sortase c1
|
| 82 | d1kshb_ |
|
not modelled |
14.5 |
5 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:RhoGDI-like |
| 83 | c2l66B_ |
|
not modelled |
14.4 |
15 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, abrb family;
PDBTitle: the dna-recognition fold of sso7c4 suggests a new member of spovt-abrb2 superfamily from archaea.
|
| 84 | d1seta1 |
|
not modelled |
13.6 |
16 |
Fold:Long alpha-hairpin
Superfamily:tRNA-binding arm
Family:Seryl-tRNA synthetase (SerRS) |
| 85 | d1qbea_ |
|
not modelled |
13.6 |
22 |
Fold:RNA bacteriophage capsid protein
Superfamily:RNA bacteriophage capsid protein
Family:RNA bacteriophage capsid protein |
| 86 | d1kwaa_ |
|
not modelled |
13.3 |
17 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:PDZ domain |
| 87 | d1ogya1 |
|
not modelled |
13.0 |
16 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 88 | c2fug4_ |
|
not modelled |
12.8 |
10 |
PDB header:oxidoreductase
Chain: 4: PDB Molecule:nadh-quinone oxidoreductase chain 4;
PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
|
| 89 | d2fug41 |
|
not modelled |
12.8 |
10 |
Fold:HydB/Nqo4-like
Superfamily:HydB/Nqo4-like
Family:Nqo4-like |
| 90 | d1gawa1 |
|
not modelled |
12.7 |
33 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 91 | d1qfja1 |
|
not modelled |
12.7 |
16 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 92 | d1q1ca1 |
|
not modelled |
12.2 |
16 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 93 | c2w1jB_ |
|
not modelled |
12.0 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:putative sortase;
PDBTitle: crystal structure of sortase c-1 (srtc-1) from2 streptococcus pneumoniae
|
| 94 | c2d90A_ |
|
not modelled |
11.9 |
18 |
PDB header:protein binding
Chain: A: PDB Molecule:pdz domain containing protein 1;
PDBTitle: solution structure of the third pdz domain of pdz domain2 containing protein 1
|
| 95 | c2xzrA_ |
|
not modelled |
11.4 |
32 |
PDB header:cell adhesion
Chain: A: PDB Molecule:immunoglobulin-binding protein eibd;
PDBTitle: escherichia coli immunoglobulin-binding protein eibd 391-438 fused2 to gcn4 adaptors
|
| 96 | d1vlfm1 |
|
not modelled |
11.4 |
17 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 97 | d1a8pa1 |
|
not modelled |
11.3 |
18 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 98 | d2bmwa1 |
|
not modelled |
11.2 |
30 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 99 | d1nbwa1 |
|
not modelled |
10.6 |
62 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Swiveling domain of dehydratase reactivase alpha subunit
Family:Swiveling domain of dehydratase reactivase alpha subunit |