1 c2qdzA_
100.0
15
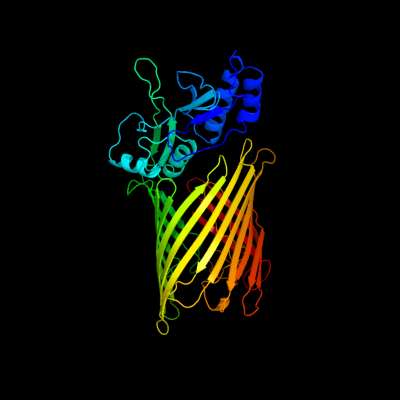
PDB header: protein transportChain: A: PDB Molecule: tpsb transporter fhac;PDBTitle: structure of the membrane protein fhac: a member of the2 omp85/tpsb transporter family
2 c3efcA_
100.0
14
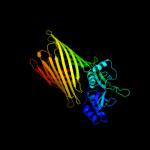
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein assembly factor yaet;PDBTitle: crystal structure of yaet periplasmic domain
3 c2x8xX_
100.0
18
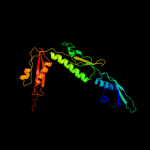
PDB header: chaperoneChain: X: PDB Molecule: tlr1789 protein;PDBTitle: structure of the n-terminal domain of omp85 from the2 thermophilic cyanobacterium thermosynechococcus elongatus
4 c3mc8A_
100.0
17
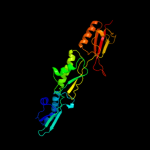
PDB header: membrane proteinChain: A: PDB Molecule: alr2269 protein;PDBTitle: potra1-3 of the periplasmic domain of omp85 from anabaena
5 c2qczA_
100.0
16
PDB header: membrane protein, protein transportChain: A: PDB Molecule: outer membrane protein assembly factor yaet;PDBTitle: structure of n-terminal domain of e. coli yaet
6 c3og5A_
99.9
16
PDB header: protein bindingChain: A: PDB Molecule: outer membrane protein assembly complex, yaet protein;PDBTitle: crystal structure of bama potra45 tandem
7 c2v9hA_
99.4
14
PDB header: protein-bindingChain: A: PDB Molecule: outer membrane protein assembly factor yaet;PDBTitle: solution structure of an escherichia coli yaet tandem potra2 domain
8 c2vh2A_
90.6
10
PDB header: cell cycleChain: A: PDB Molecule: cell division protein ftsq;PDBTitle: crystal structure of cell divison protein ftsq from2 yersinia enterecolitica
9 d1vqza1
89.2
14
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: SP1160 C-terminal domain-like10 c1vqzA_
87.4
11
PDB header: ligaseChain: A: PDB Molecule: lipoate-protein ligase, putative;PDBTitle: crystal structure of a putative lipoate-protein ligase a (sp_1160)2 from streptococcus pneumoniae tigr4 at 1.99 a resolution
11 c2vh1A_
78.6
14
PDB header: cell cycleChain: A: PDB Molecule: cell division protein ftsq;PDBTitle: crystal structure of bacterial cell division protein ftsq2 from e.coli
12 d1x2ga1
72.6
15
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: SP1160 C-terminal domain-like13 c1x2gB_
42.2
14
PDB header: ligaseChain: B: PDB Molecule: lipoate-protein ligase a;PDBTitle: crystal structure of lipate-protein ligase a from2 escherichia coli
14 c2k4vA_
29.9
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pa1076;PDBTitle: solution structure of uncharacterized protein pa1076 from2 pseudomonas aeruginosa. northeast structural genomics3 consortium (nesg) target pat3, ontario center for4 structural proteomics target pa1076 .
15 d2h7aa1
17.8
17
Fold: YcgL-likeSuperfamily: YcgL-likeFamily: YcgL-like16 c3e19D_
14.6
10
PDB header: transcription regulator, metal binding pChain: D: PDB Molecule: feoa;PDBTitle: crystal structure of iron uptake regulatory protein (feoa) solved by2 sulfur sad in a monoclinic space group
17 d1p4ta_
13.6
6
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein18 d1m6ya1
13.3
25
Fold: SAM domain-likeSuperfamily: Putative methyltransferase TM0872, insert domainFamily: Putative methyltransferase TM0872, insert domain19 d2fnaa1
9.4
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Helicase DNA-binding domain20 d1nija2
9.1
14
Fold: Hypothetical protein YjiA, C-terminal domainSuperfamily: Hypothetical protein YjiA, C-terminal domainFamily: Hypothetical protein YjiA, C-terminal domain21 d2gcxa1
not modelled
9.1
8
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: FeoA-like22 c2y50A_
not modelled
8.8
19
PDB header: hydrolaseChain: A: PDB Molecule: collagenase;PDBTitle: crystal structure of collagenase g from clostridium2 histolyticum at 2.80 angstrom resolution
23 d2fsqa1
not modelled
8.4
22
Fold: LigT-likeSuperfamily: LigT-likeFamily: Atu0111-like24 d1vqqa1
not modelled
7.8
9
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Penicillin binding protein 2a (PBP2A), N-terminal domain25 c2k0lA_
not modelled
7.7
14
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr structure of the transmembrane domain of the outer2 membrane protein a from klebsiella pneumoniae in dhpc3 micelles.
26 c3hrlA_
not modelled
7.4
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: endonuclease-like protein;PDBTitle: crystal structure of a putative endonuclease-like protein (ngo0050)2 from neisseria gonorrhoeae
27 d1ssna_
not modelled
7.4
17
Fold: beta-Grasp (ubiquitin-like)Superfamily: Staphylokinase/streptokinaseFamily: Staphylokinase/streptokinase28 d1wg8a1
not modelled
7.4
32
Fold: SAM domain-likeSuperfamily: Putative methyltransferase TM0872, insert domainFamily: Putative methyltransferase TM0872, insert domain29 c3bd0D_
not modelled
7.3
17
PDB header: peptide binding proteinChain: D: PDB Molecule: protein memo1;PDBTitle: crystal structure of memo, form ii
30 c1bmlD_
not modelled
7.3
14
PDB header: blood clottingChain: D: PDB Molecule: streptokinase;PDBTitle: complex of the catalytic domain of human plasmin and streptokinase
31 d1g90a_
not modelled
6.0
12
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein32 d1seia_
not modelled
6.0
23
Fold: Ribosomal protein S8Superfamily: Ribosomal protein S8Family: Ribosomal protein S833 c3kvnA_
not modelled
5.7
10
PDB header: hydrolaseChain: A: PDB Molecule: esterase esta;PDBTitle: crystal structure of the full-length autotransporter esta from2 pseudomonas aeruginosa
34 c3fpnB_
not modelled
5.6
16
PDB header: dna binding proteinChain: B: PDB Molecule: geobacillus stearothermophilus uvrb interactionPDBTitle: crystal structure of uvra-uvrb interaction domains
35 c1nijA_
not modelled
5.2
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yjia;PDBTitle: yjia protein