1 c3giaA_
100.0
17
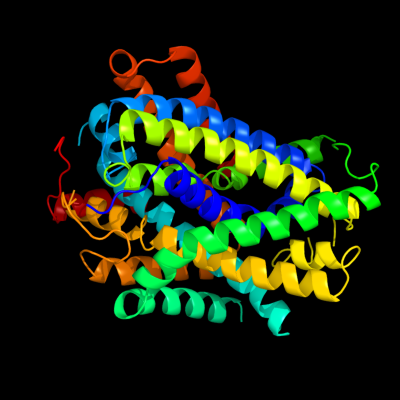
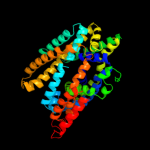
PDB header: transport proteinChain: A: PDB Molecule: uncharacterized protein mj0609;PDBTitle: crystal structure of apct transporter
2 c3lrcC_
100.0
21
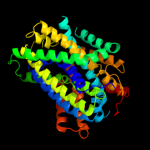
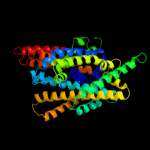
PDB header: transport proteinChain: C: PDB Molecule: arginine/agmatine antiporter;PDBTitle: structure of e. coli adic (p1)
3 c2jlnA_
99.9
9
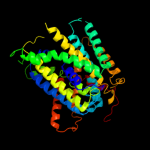
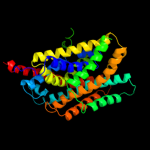
PDB header: membrane proteinChain: A: PDB Molecule: mhp1;PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
4 c2xq2A_
99.1
13
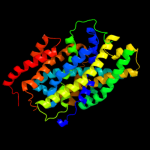
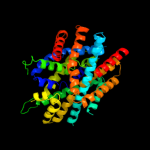
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: structure of the k294a mutant of vsglt
5 c3dh4A_
98.9
12
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
6 c2w8aC_
97.0
16
PDB header: membrane proteinChain: C: PDB Molecule: glycine betaine transporter betp;PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
7 d2a65a1
96.6
13
Fold: SNF-likeSuperfamily: SNF-likeFamily: SNF-like8 c3hfxA_
95.2
11
PDB header: transport proteinChain: A: PDB Molecule: l-carnitine/gamma-butyrobetaine antiporter;PDBTitle: crystal structure of carnitine transporter
9 c2kncA_
15.0
24
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
10 c1m57H_
13.2
17
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c oxidase;PDBTitle: structure of cytochrome c oxidase from rhodobacter2 sphaeroides (eq(i-286) mutant))
11 d1fftc_
10.5
13
Fold: Cytochrome c oxidase subunit III-likeSuperfamily: Cytochrome c oxidase subunit III-likeFamily: Cytochrome c oxidase subunit III-like12 d1fftb2
10.2
9
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region13 c1qleB_
9.9
10
PDB header: oxidoreductase/immune systemChain: B: PDB Molecule: cytochrome c oxidase polypeptide ii;PDBTitle: cryo-structure of the paracoccus denitrificans four-subunit2 cytochrome c oxidase in the completely oxidized state3 complexed with an antibody fv fragment
14 d3ehbb2
9.4
20
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region15 c2k9yA_
9.4
28
PDB header: transferaseChain: A: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
16 c2k9yB_
9.4
28
PDB header: transferaseChain: B: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
17 c2k1aA_
9.1
26
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
18 d1dt9a3
9.0
50
Fold: N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1Superfamily: N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1Family: N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF119 c2l1uA_
8.9
22
PDB header: oxidoreductaseChain: A: PDB Molecule: methionine-r-sulfoxide reductase b2, mitochondrial;PDBTitle: structure-functional analysis of mammalian msrb2 protein
20 c3b71E_
8.7
44
PDB header: protein bindingChain: E: PDB Molecule: t-cell surface glycoprotein cd4;PDBTitle: cd4 endocytosis motif bound to the focal adhesion targeting (fat)2 domain of the focal adhesion kinase
21 c3b71D_
not modelled
8.7
44
PDB header: protein bindingChain: D: PDB Molecule: t-cell surface glycoprotein cd4;PDBTitle: cd4 endocytosis motif bound to the focal adhesion targeting (fat)2 domain of the focal adhesion kinase
22 c3rceA_
not modelled
8.5
19
PDB header: transferase/peptideChain: A: PDB Molecule: oligosaccharide transferase to n-glycosylate proteins;PDBTitle: bacterial oligosaccharyltransferase pglb
23 d3dtub2
not modelled
8.3
20
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region24 c3qe7A_
not modelled
8.3
13
PDB header: transport proteinChain: A: PDB Molecule: uracil permease;PDBTitle: crystal structure of uracil transporter--uraa
25 c3e20C_
not modelled
8.0
40
PDB header: translationChain: C: PDB Molecule: eukaryotic peptide chain release factor subunit 1;PDBTitle: crystal structure of s.pombe erf1/erf3 complex
26 c2kfeA_
not modelled
7.9
56
PDB header: antimicrobial proteinChain: A: PDB Molecule: meucin-24;PDBTitle: solution structure of meucin-24
27 d1xg8a_
not modelled
7.7
5
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: YuzD-like28 d2obpa1
not modelled
7.7
31
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ReutB4095-like29 c2k21A_
not modelled
7.4
10
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily ePDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
30 d1t98a2
not modelled
7.2
8
Fold: STAT-likeSuperfamily: MukF C-terminal domain-likeFamily: MukF C-terminal domain-like31 c1ar1B_
not modelled
7.1
20
PDB header: complex (oxidoreductase/antibody)Chain: B: PDB Molecule: cytochrome c oxidase;PDBTitle: structure at 2.7 angstrom resolution of the paracoccus2 denitrificans two-subunit cytochrome c oxidase complexed3 with an antibody fv fragment
32 d2cmyb1
not modelled
7.1
29
Fold: Toxic hairpinSuperfamily: VhTI-likeFamily: VhTI-like33 c2cmyB_
not modelled
7.1
29
PDB header: hydrolaseChain: B: PDB Molecule: veronica hederifolia trypsin inhibitor;PDBTitle: crystal complex between bovine trypsin and veronica2 hederifolia trypsin inhibitor
34 d1vc4a_
not modelled
6.9
40
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes35 c1ujlA_
not modelled
6.8
11
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily hPDBTitle: solution structure of the herg k+ channel s5-p2 extracellular linker
36 d1xmec1
not modelled
6.6
23
Fold: Single transmembrane helixSuperfamily: Bacterial ba3 type cytochrome c oxidase subunit IIaFamily: Bacterial ba3 type cytochrome c oxidase subunit IIa37 d1ic8a2
not modelled
6.2
15
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: POU-specific domain38 c3dwuA_
not modelled
6.1
60
PDB header: biosynthetic proteinChain: A: PDB Molecule: elongation factor tu-b;PDBTitle: transition-state model conformation of the switch i region2 fitted into the cryo-em map of the eef2.80s.alf4.gdp3 complex
39 d1a53a_
not modelled
6.0
40
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes40 d1iwpa_
not modelled
6.0
44
Fold: TIM beta/alpha-barrelSuperfamily: Cobalamin (vitamin B12)-dependent enzymesFamily: Diol dehydratase, alpha subunit41 d1eexa_
not modelled
6.0
33
Fold: TIM beta/alpha-barrelSuperfamily: Cobalamin (vitamin B12)-dependent enzymesFamily: Diol dehydratase, alpha subunit42 c2cpbA_
not modelled
5.9
12
PDB header: viral proteinChain: A: PDB Molecule: m13 major coat protein;PDBTitle: solution nmr structures of the major coat protein of2 filamentous bacteriophage m13 solubilized in3 dodecylphosphocholine micelles, 25 lowest energy structures
43 c1t0fC_
not modelled
5.8
20
PDB header: dna binding proteinChain: C: PDB Molecule: transposon tn7 transposition protein tnsc;PDBTitle: crystal structure of the tnsa/tnsc(504-555) complex
44 c2rddB_
not modelled
5.7
10
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
45 d1efea_
not modelled
5.6
20
Fold: Insulin-likeSuperfamily: Insulin-likeFamily: Insulin-like46 d2iuba2
not modelled
5.6
20
Fold: Transmembrane helix hairpinSuperfamily: Magnesium transport protein CorA, transmembrane regionFamily: Magnesium transport protein CorA, transmembrane region47 d1joya_
not modelled
5.5
15
Fold: ROP-likeSuperfamily: Homodimeric domain of signal transducing histidine kinaseFamily: Homodimeric domain of signal transducing histidine kinase48 c1unhD_
not modelled
5.5
11
PDB header: cell cycleChain: D: PDB Molecule: cyclin-dependent kinase 5 activator 1;PDBTitle: structural mechanism for the inhibition of cdk5-p25 by2 roscovitine, aloisine and indirubin.
49 d1lr7a1
not modelled
5.4
12
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: EGF/LamininFamily: Follistatin (FS) module N-terminal domain, FS-N50 d1unld_
not modelled
5.4
11
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Cyclin51 d2nr9a1
not modelled
5.4
9
Fold: Rhomboid-likeSuperfamily: Rhomboid-likeFamily: Rhomboid-like52 d1eah1_
not modelled
5.3
57
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Picornaviridae-like VP (VP1, VP2, VP3 and VP4)53 d1pmxa_
not modelled
5.2
40
Fold: Insulin-likeSuperfamily: Insulin-likeFamily: Insulin-like54 d1j75a_
not modelled
5.2
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Z-DNA binding domain55 c2qtyB_
not modelled
5.1
23
PDB header: hydrolaseChain: B: PDB Molecule: poly(adp-ribose) glycohydrolase arh3;PDBTitle: crystal structure of mouse adp-ribosylhydrolase 3 (marh3)