| 1 |
|
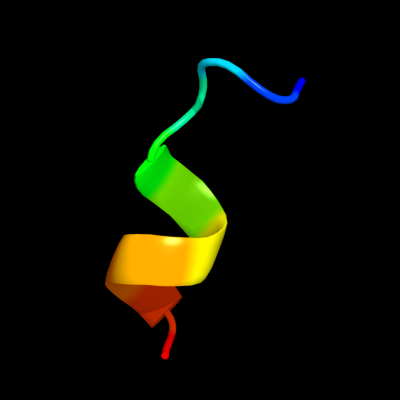
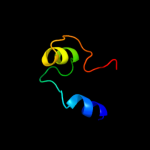
PDB 2zxx chain A
Region: 84 - 94
Aligned: 11
Modelled: 11
Confidence: 31.0%
Identity: 36%
PDB header:cell cycle/replication
Chain: A: PDB Molecule:geminin;
PDBTitle: crystal structure of cdt1/geminin complex
Phyre2
| 2 |
|
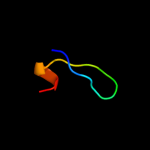
PDB 1i5p chain A domain 3
Region: 49 - 93
Aligned: 39
Modelled: 45
Confidence: 22.3%
Identity: 21%
Fold: Toxins' membrane translocation domains
Superfamily: delta-Endotoxin (insectocide), N-terminal domain
Family: delta-Endotoxin (insectocide), N-terminal domain
Phyre2
| 3 |
|
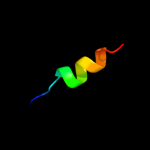
PDB 2wvr chain B
Region: 84 - 94
Aligned: 11
Modelled: 11
Confidence: 19.2%
Identity: 36%
PDB header:replication
Chain: B: PDB Molecule:geminin;
PDBTitle: human cdt1:geminin complex
Phyre2
| 4 |
|
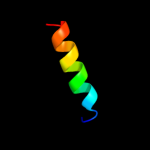
PDB 1fo8 chain A
Region: 87 - 125
Aligned: 29
Modelled: 39
Confidence: 10.0%
Identity: 28%
Fold: Nucleotide-diphospho-sugar transferases
Superfamily: Nucleotide-diphospho-sugar transferases
Family: N-acetylglucosaminyltransferase I
Phyre2
| 5 |
|
PDB 1iij chain A
Region: 41 - 60
Aligned: 20
Modelled: 20
Confidence: 9.8%
Identity: 30%
PDB header:signaling protein
Chain: A: PDB Molecule:erbb-2 receptor protein-tyrosine kinase;
PDBTitle: solution structure of the neu/erbb-2 membrane spanning2 segment
Phyre2
| 6 |
|
PDB 1z65 chain A
Region: 7 - 18
Aligned: 12
Modelled: 12
Confidence: 9.6%
Identity: 42%
PDB header:unknown function
Chain: A: PDB Molecule:prion-like protein doppel;
PDBTitle: mouse doppel 1-30 peptide
Phyre2
| 7 |
|
PDB 1x4l chain A domain 2
Region: 53 - 68
Aligned: 16
Modelled: 16
Confidence: 7.6%
Identity: 31%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 8 |
|
PDB 1ffg chain B
Region: 9 - 15
Aligned: 7
Modelled: 7
Confidence: 7.3%
Identity: 57%
Fold: Ferredoxin-like
Superfamily: CheY-binding domain of CheA
Family: CheY-binding domain of CheA
Phyre2
| 9 |
|
PDB 1eys chain H domain 2
Region: 110 - 124
Aligned: 15
Modelled: 15
Confidence: 7.2%
Identity: 20%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction centre subunit H, transmembrane region
Family: Photosystem II reaction centre subunit H, transmembrane region
Phyre2
| 10 |
|
PDB 1a0o chain H
Region: 9 - 15
Aligned: 7
Modelled: 7
Confidence: 7.2%
Identity: 57%
PDB header:chemotaxis
Chain: H: PDB Molecule:chea;
PDBTitle: chey-binding domain of chea in complex with chey
Phyre2
| 11 |
|
PDB 1q3j chain A
Region: 48 - 58
Aligned: 11
Modelled: 11
Confidence: 5.8%
Identity: 36%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: Gurmarin-like
Family: Antifungal peptide
Phyre2
| 12 |
|
PDB 1bhb chain A
Region: 38 - 50
Aligned: 13
Modelled: 13
Confidence: 5.8%
Identity: 31%
PDB header:photoreceptor
Chain: A: PDB Molecule:bacteriorhodopsin;
PDBTitle: three-dimensional structure of (1-71) bacterioopsin2 solubilized in methanol-chloroform and sds micelles3 determined by 15n-1h heteronuclear nmr spectroscopy
Phyre2
| 13 |
|
PDB 3t7h chain B
Region: 86 - 94
Aligned: 9
Modelled: 9
Confidence: 5.7%
Identity: 56%
PDB header:ligase
Chain: B: PDB Molecule:ubiquitin-like modifier-activating enzyme atg7;
PDBTitle: atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and2 mechanism in the autophagy pathway
Phyre2
| 14 |
|
PDB 3hls chain E
Region: 42 - 52
Aligned: 11
Modelled: 11
Confidence: 5.2%
Identity: 55%
PDB header:signaling protein
Chain: E: PDB Molecule:guanylate cyclase soluble subunit beta-1;
PDBTitle: crystal structure of the signaling helix coiled-coil doimain2 of the beta-1 subunit of the soluble guanylyl cyclase
Phyre2
| 15 |
|
PDB 3n0a chain A
Region: 1 - 18
Aligned: 18
Modelled: 18
Confidence: 5.2%
Identity: 6%
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase auxilin;
PDBTitle: crystal structure of auxilin (40-400)
Phyre2