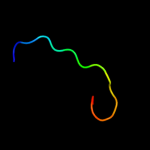
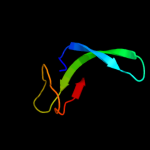
1 d2hqha1
22.3
41
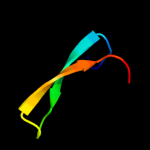
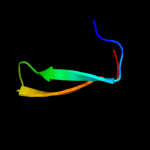
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain2 d2coya1
22.3
33
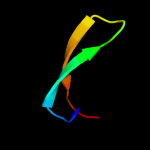
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain3 d1w4xa2
19.0
53
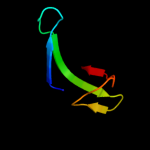
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains4 c3iz6D_
16.3
45
PDB header: ribosomeChain: D: PDB Molecule: 40s ribosomal protein s4 (s4e);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
5 c2xzmW_
16.1
27
PDB header: ribosomeChain: W: PDB Molecule: 40s ribosomal protein s4;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
6 c3uvzB_
16.1
29
PDB header: lyaseChain: B: PDB Molecule: phosphoribosylaminoimidazole carboxylase, atpase subunit;PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase, atpase2 subunit from burkholderia ambifaria
7 d1q1ra2
15.0
25
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains8 d1g7sa2
14.4
23
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors9 c3kbgA_
13.8
27
PDB header: ribosomal proteinChain: A: PDB Molecule: 30s ribosomal protein s4e;PDBTitle: crystal structure of the 30s ribosomal protein s4e from2 thermoplasma acidophilum. northeast structural genomics3 consortium target tar28.
10 d1zunb1
13.6
24
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors11 c3orqA_
13.3
30
PDB header: ligase,biosynthetic proteinChain: A: PDB Molecule: n5-carboxyaminoimidazole ribonucleotide synthetase;PDBTitle: crystal structure of n5-carboxyaminoimidazole synthetase from2 staphylococcus aureus complexed with adp
12 d1whka_
11.8
14
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain13 c2zc6A_
11.4
36
PDB header: biosynthetic proteinChain: A: PDB Molecule: penicillin-binding protein 1a;PDBTitle: penicillin-binding protein 1a (pbp 1a) acyl-enzyme complex2 (tebipenem) from streptococcus pneumoniae
14 c2zc6C_
11.4
36
PDB header: biosynthetic proteinChain: C: PDB Molecule: penicillin-binding protein 1a;PDBTitle: penicillin-binding protein 1a (pbp 1a) acyl-enzyme complex2 (tebipenem) from streptococcus pneumoniae
15 c2zc5C_
11.4
36
PDB header: biosynthetic proteinChain: C: PDB Molecule: penicillin-binding protein 1a;PDBTitle: penicillin-binding protein 1a (pbp 1a) acyl-enzyme complex2 (biapenem) from streptococcus pneumoniae
16 c2zc5A_
11.4
36
PDB header: biosynthetic proteinChain: A: PDB Molecule: penicillin-binding protein 1a;PDBTitle: penicillin-binding protein 1a (pbp 1a) acyl-enzyme complex2 (biapenem) from streptococcus pneumoniae
17 c2v2fA_
11.4
36
PDB header: transferaseChain: A: PDB Molecule: penicillin binding protein 1a;PDBTitle: crystal structure of pbp1a from drug-resistant strain 52042 from streptococcus pneumoniae
18 d1li4a1
10.5
29
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain19 d1l7da1
9.5
43
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain20 c3epmB_
9.2
38
PDB header: biosynthetic proteinChain: B: PDB Molecule: thiamine biosynthesis protein thic;PDBTitle: crystal structure of caulobacter crescentus thic
21 c2d7gD_
not modelled
8.4
22
PDB header: hydrolaseChain: D: PDB Molecule: primosomal protein n;PDBTitle: crystal structure of the aa complex of the n-terminal2 domain of pria
22 c3gvpB_
not modelled
8.2
26
PDB header: hydrolaseChain: B: PDB Molecule: adenosylhomocysteinase 3;PDBTitle: human sahh-like domain of human adenosylhomocysteinase 3
23 d1stma_
not modelled
8.1
38
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Satellite virusesFamily: Satellite viruses24 c3q2oB_
not modelled
7.8
26
PDB header: lyaseChain: B: PDB Molecule: phosphoribosylaminoimidazole carboxylase, atpase subunit;PDBTitle: crystal structure of purk: n5-carboxyaminoimidazole ribonucleotide2 synthetase
25 d1d7ya2
not modelled
7.5
25
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains26 d1fcda1
not modelled
7.3
40
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains27 d1wb1a1
not modelled
7.3
18
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors28 d1g47a1
not modelled
7.1
36
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain29 d1muga_
not modelled
6.8
33
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Mug-like30 c2xz2A_
not modelled
6.6
21
PDB header: rna-binding proteinChain: A: PDB Molecule: cstf-50, isoform b;PDBTitle: crystal structure of cstf-50 homodimerization domain
31 d1d1ta2
not modelled
6.5
31
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain32 c1zfnA_
not modelled
6.2
35
PDB header: transferaseChain: A: PDB Molecule: adenylyltransferase thif;PDBTitle: structural analysis of escherichia coli thif
33 d1pjqa1
not modelled
6.2
12
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Siroheme synthase N-terminal domain-like34 c2yyyB_
not modelled
6.1
36
PDB header: oxidoreductaseChain: B: PDB Molecule: glyceraldehyde-3-phosphate dehydrogenase;PDBTitle: crystal structure of glyceraldehyde-3-phosphate2 dehydrogenase
35 c2gzaB_
not modelled
6.1
31
PDB header: hydrolaseChain: B: PDB Molecule: type iv secretion system protein virb11;PDBTitle: crystal structure of the virb11 atpase from the brucella suis type iv2 secretion system in complex with sulphate
36 d2czca2
not modelled
6.1
45
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain37 d1kola2
not modelled
6.0
35
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain38 d1v8ba1
not modelled
5.8
29
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain39 c1cf2Q_
not modelled
5.7
33
PDB header: oxidoreductaseChain: Q: PDB Molecule: protein (glyceraldehyde-3-phosphatePDBTitle: three-dimensional structure of d-glyceraldehyde-3-phosphate2 dehydrogenase from the hyperthermophilic archaeon3 methanothermus fervidus
40 c1gpjA_
not modelled
5.5
30
PDB header: reductaseChain: A: PDB Molecule: glutamyl-trna reductase;PDBTitle: glutamyl-trna reductase from methanopyrus kandleri
41 c3s5wB_
not modelled
5.5
8
PDB header: oxidoreductaseChain: B: PDB Molecule: l-ornithine 5-monooxygenase;PDBTitle: ornithine hydroxylase (pvda) from pseudomonas aeruginosa
42 d1gpja2
not modelled
5.5
38
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain43 d1pyya3
not modelled
5.3
50
Fold: Penicillin binding protein dimerisation domainSuperfamily: Penicillin binding protein dimerisation domainFamily: Penicillin binding protein dimerisation domain44 c1w4xA_
not modelled
5.3
25
PDB header: oxygenaseChain: A: PDB Molecule: phenylacetone monooxygenase;PDBTitle: phenylacetone monooxygenase, a baeyer-villiger2 monooxygenase
45 d1dssg1
not modelled
5.3
33
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain46 d1leha1
not modelled
5.1
36
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain