| 1 |
|
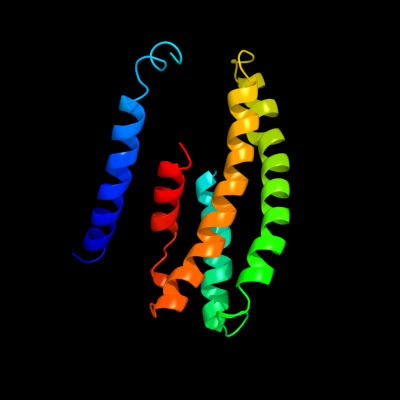
PDB 2yvx chain D
Region: 25 - 190
Aligned: 143
Modelled: 144
Confidence: 89.6%
Identity: 17%
PDB header:transport protein
Chain: D: PDB Molecule:mg2+ transporter mgte;
PDBTitle: crystal structure of magnesium transporter mgte
Phyre2
| 2 |
|
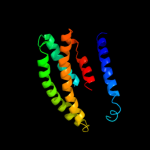
PDB 2yvx chain A domain 3
Region: 25 - 190
Aligned: 143
Modelled: 144
Confidence: 44.9%
Identity: 17%
Fold: MgtE membrane domain-like
Superfamily: MgtE membrane domain-like
Family: MgtE membrane domain-like
Phyre2
| 3 |
|
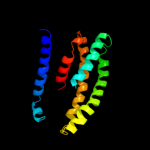
PDB 1l7v chain A
Region: 20 - 200
Aligned: 162
Modelled: 162
Confidence: 16.2%
Identity: 16%
Fold: ABC transporter involved in vitamin B12 uptake, BtuC
Superfamily: ABC transporter involved in vitamin B12 uptake, BtuC
Family: ABC transporter involved in vitamin B12 uptake, BtuC
Phyre2
| 4 |
|
PDB 1z0k chain B domain 1
Region: 76 - 89
Aligned: 14
Modelled: 14
Confidence: 13.7%
Identity: 36%
Fold: Long alpha-hairpin
Superfamily: Rabenosyn-5 Rab-binding domain-like
Family: Rabenosyn-5 Rab-binding domain-like
Phyre2
| 5 |
|
PDB 3zv0 chain A
Region: 72 - 113
Aligned: 42
Modelled: 42
Confidence: 13.3%
Identity: 19%
PDB header:cell cycle
Chain: A: PDB Molecule:protein shq1;
PDBTitle: structure of the shq1p-cbf5p complex
Phyre2
| 6 |
|
PDB 3rfu chain C
Region: 23 - 108
Aligned: 86
Modelled: 86
Confidence: 13.1%
Identity: 17%
PDB header:hydrolase, membrane protein
Chain: C: PDB Molecule:copper efflux atpase;
PDBTitle: crystal structure of a copper-transporting pib-type atpase
Phyre2
| 7 |
|
PDB 1yzm chain A domain 1
Region: 76 - 89
Aligned: 14
Modelled: 14
Confidence: 12.8%
Identity: 36%
Fold: Long alpha-hairpin
Superfamily: Rabenosyn-5 Rab-binding domain-like
Family: Rabenosyn-5 Rab-binding domain-like
Phyre2
| 8 |
|
PDB 1ckx chain A
Region: 46 - 56
Aligned: 11
Modelled: 11
Confidence: 11.8%
Identity: 45%
PDB header:metal transport
Chain: A: PDB Molecule:cystic fibrosis transmembrane conductance
PDBTitle: cystic fibrosis transmembrane conductance regulator:2 solution structures of peptides based on the phe508 region,3 the most common site of disease-causing delta-f508 mutation
Phyre2
| 9 |
|
PDB 1cii chain A
Region: 122 - 167
Aligned: 46
Modelled: 46
Confidence: 9.6%
Identity: 15%
PDB header:transmembrane protein
Chain: A: PDB Molecule:colicin ia;
PDBTitle: colicin ia
Phyre2
| 10 |
|
PDB 2ks1 chain B
Region: 58 - 84
Aligned: 27
Modelled: 27
Confidence: 8.6%
Identity: 30%
PDB header:transferase
Chain: B: PDB Molecule:epidermal growth factor receptor;
PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
Phyre2
| 11 |
|
PDB 3p5n chain A
Region: 5 - 87
Aligned: 83
Modelled: 83
Confidence: 8.1%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:riboflavin uptake protein;
PDBTitle: structure and mechanism of the s component of a bacterial ecf2 transporter
Phyre2
| 12 |
|
PDB 2akz chain A domain 2
Region: 72 - 87
Aligned: 16
Modelled: 16
Confidence: 8.0%
Identity: 25%
Fold: Enolase N-terminal domain-like
Superfamily: Enolase N-terminal domain-like
Family: Enolase N-terminal domain-like
Phyre2
| 13 |
|
PDB 1ckw chain A
Region: 47 - 56
Aligned: 10
Modelled: 10
Confidence: 7.2%
Identity: 50%
PDB header:metal transport
Chain: A: PDB Molecule:protein (cystic fibrosis transmembrane
PDBTitle: cystic fibrosis transmembrane conductance regulator:2 solution structures of peptides based on the phe508 region,3 the most common site of disease-causing delta-f508 mutation
Phyre2
| 14 |
|
PDB 1uny chain A
Region: 161 - 178
Aligned: 18
Modelled: 18
Confidence: 6.6%
Identity: 28%
PDB header:four helix bundle
Chain: A: PDB Molecule:general control protein gcn4;
PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
Phyre2
| 15 |
|
PDB 2ju0 chain B
Region: 15 - 33
Aligned: 19
Modelled: 19
Confidence: 5.3%
Identity: 26%
PDB header:metal binding protein/signaling protein
Chain: B: PDB Molecule:phosphatidylinositol 4-kinase pik1;
PDBTitle: structure of yeast frequenin bound to pdtins 4-kinase
Phyre2