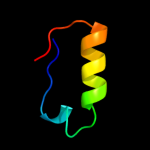
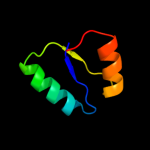
1 d1xn7a_
58.5
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Hypothetical protein YhgG2 d1k8kg_
58.1
23
Fold: alpha-alpha superhelixSuperfamily: Arp2/3 complex 16 kDa subunit ARPC5Family: Arp2/3 complex 16 kDa subunit ARPC53 d1ylfa1
48.9
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Transcriptional regulator Rrf24 c3k69A_
44.5
17
PDB header: transcriptionChain: A: PDB Molecule: putative transcription regulator;PDBTitle: crystal structure of a putative transcriptional regulator (lp_0360)2 from lactobacillus plantarum at 1.95 a resolution
5 d1vcta1
41.2
8
Fold: Spectrin repeat-likeSuperfamily: PhoU-likeFamily: PhoU-like6 d1xqoa_
40.8
11
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: AgoG-like7 d2ihta1
40.5
55
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain8 c3dwlG_
40.4
20
PDB header: structural proteinChain: G: PDB Molecule: actin-related protein 2/3 complex subunit 5;PDBTitle: crystal structure of fission yeast arp2/3 complex lacking the arp22 subunit
9 d1s8na_
37.1
9
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related10 d1xd7a_
33.2
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Transcriptional regulator Rrf211 d1ufma_
32.7
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PCI domain (PINT motif)12 d1t9ba1
29.9
33
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain13 d2ji7a1
29.7
21
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain14 d1q6za1
29.6
28
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain15 d1zpda1
28.0
28
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain16 d1ozha1
27.7
31
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain17 c3c3mA_
26.1
26
PDB header: signaling proteinChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of the n-terminal domain of response regulator2 receiver protein from methanoculleus marisnigri jr1
18 c2vbgB_
24.4
17
PDB header: lyaseChain: B: PDB Molecule: branched-chain alpha-ketoacid decarboxylase;PDBTitle: the complex structure of the branched-chain keto acid2 decarboxylase (kdca) from lactococcus lactis with 2r-1-3 hydroxyethyl-deazathdp
19 d1pvda1
23.2
10
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain20 d1p6qa_
23.0
13
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related21 c2y75F_
not modelled
20.9
7
PDB header: transcriptionChain: F: PDB Molecule: hth-type transcriptional regulator cymr;PDBTitle: the structure of cymr (yrzc) the global cysteine regulator2 of b. subtilis
22 d1u0sy_
not modelled
20.5
22
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related23 c3t6kB_
not modelled
20.4
20
PDB header: signaling proteinChain: B: PDB Molecule: response regulator receiver;PDBTitle: crystal structure of a hypothetical response regulator (caur_3799)2 from chloroflexus aurantiacus j-10-fl at 1.86 a resolution
24 d1jbea_
not modelled
19.1
13
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related25 c3h1gA_
not modelled
17.7
13
PDB header: signaling proteinChain: A: PDB Molecule: chemotaxis protein chey homolog;PDBTitle: crystal structure of chey mutant t84a of helicobacter pylori
26 d1ovma1
not modelled
17.2
17
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain27 c3c5tB_
not modelled
16.9
26
PDB header: signaling protein/signaling proteinChain: B: PDB Molecule: exendin-4;PDBTitle: crystal structure of the ligand-bound glucagon-like peptide-1 receptor2 extracellular domain
28 c3hzhA_
not modelled
16.8
21
PDB header: signaling proteinChain: A: PDB Molecule: chemotaxis response regulator (chey-3);PDBTitle: crystal structure of the chex-chey-bef3-mg+2 complex from2 borrelia burgdorferi
29 c3khtA_
not modelled
16.8
11
PDB header: signaling proteinChain: A: PDB Molecule: response regulator;PDBTitle: crystal structure of response regulator from hahella chejuensis
30 d1ny5a1
not modelled
16.7
9
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related31 c1ojhK_
not modelled
16.2
19
PDB header: protein bindingChain: K: PDB Molecule: nbla;PDBTitle: crystal structure of nbla from pcc 7120
32 d2pl1a1
not modelled
16.0
15
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related33 c2qxyB_
not modelled
16.0
24
PDB header: transcriptionChain: B: PDB Molecule: response regulator;PDBTitle: crystal structure of a response regulator from thermotoga2 maritima
34 c2zwmA_
not modelled
15.9
18
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein yycf;PDBTitle: crystal structure of yycf receiver domain from bacillus2 subtilis
35 c3i42A_
not modelled
15.8
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: response regulator receiver domain protein (chey-PDBTitle: structure of response regulator receiver domain (chey-like)2 from methylobacillus flagellatus
36 c1zpdA_
not modelled
15.7
28
PDB header: alcohol fermentationChain: A: PDB Molecule: pyruvate decarboxylase;PDBTitle: pyruvate decarboxylase from zymomonas mobilis
37 c3grcD_
not modelled
15.6
13
PDB header: transferaseChain: D: PDB Molecule: sensor protein, kinase;PDBTitle: crystal structure of a sensor protein from polaromonas sp.2 js666
38 d2fug21
not modelled
15.6
8
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: NQO2-like39 d1ojha_
not modelled
15.5
19
Fold: NblA-likeSuperfamily: NblA-likeFamily: NblA-like40 d1zgza1
not modelled
15.3
11
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related41 c3txmA_
not modelled
15.3
18
PDB header: hydrolase, protein bindingChain: A: PDB Molecule: 26s proteasome regulatory complex subunit p42b;PDBTitle: crystal structure of rpn6 from drosophila melanogaster, gd(3+) complex
42 c3cg4A_
not modelled
15.0
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: response regulator receiver domain protein (chey-like);PDBTitle: crystal structure of response regulator receiver domain protein (chey-2 like) from methanospirillum hungatei jf-1
43 c3c97A_
not modelled
14.8
13
PDB header: signaling protein, transferaseChain: A: PDB Molecule: signal transduction histidine kinase;PDBTitle: crystal structure of the response regulator receiver domain2 of a signal transduction histidine kinase from aspergillus3 oryzae
44 c3hv2B_
not modelled
13.5
22
PDB header: signaling proteinChain: B: PDB Molecule: response regulator/hd domain protein;PDBTitle: crystal structure of signal receiver domain of hd domain-2 containing protein from pseudomonas fluorescens pf-5
45 d1mvoa_
not modelled
13.4
22
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related46 c3cf4G_
not modelled
13.3
21
PDB header: oxidoreductaseChain: G: PDB Molecule: acetyl-coa decarboxylase/synthase epsilon subunit;PDBTitle: structure of the codh component of the m. barkeri acds complex
47 d1ybha1
not modelled
13.3
30
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain48 c1jrjA_
not modelled
13.1
29
PDB header: hormone/growth factorChain: A: PDB Molecule: exendin-4;PDBTitle: solution structure of exendin-4 in 30-vol% trifluoroethanol
49 d1ig6a_
not modelled
13.0
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: ARID-likeFamily: ARID domain50 c2j48A_
not modelled
12.3
18
PDB header: transferaseChain: A: PDB Molecule: two-component sensor kinase;PDBTitle: nmr structure of the pseudo-receiver domain of the cika2 protein.
51 c3a0rB_
not modelled
12.2
18
PDB header: transferaseChain: B: PDB Molecule: response regulator;PDBTitle: crystal structure of histidine kinase thka (tm1359) in complex with2 response regulator protein trra (tm1360)
52 d1kgsa2
not modelled
12.1
7
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related53 c1ozhD_
not modelled
12.0
31
PDB header: lyaseChain: D: PDB Molecule: acetolactate synthase, catabolic;PDBTitle: the crystal structure of klebsiella pneumoniae acetolactate2 synthase with enzyme-bound cofactor and with an unusual3 intermediate.
54 d2ayxa1
not modelled
11.7
20
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related55 d1bgpa_
not modelled
11.6
26
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: CCP-like56 d1ys7a2
not modelled
11.5
23
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related57 c1d0rA_
not modelled
10.9
22
PDB header: hormone/growth factorChain: A: PDB Molecule: glucagon-like peptide-1-(7-36)-amide;PDBTitle: solution structure of glucagon-like peptide-1-(7-36)-amide2 in trifluoroethanol/water
58 d2ez9a1
not modelled
10.9
25
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain59 c3cixA_
not modelled
10.8
26
PDB header: adomet binding proteinChain: A: PDB Molecule: fefe-hydrogenase maturase;PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
60 d1yioa2
not modelled
10.6
16
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related61 c2rohA_
not modelled
10.4
18
PDB header: dna binding proteinChain: A: PDB Molecule: telomere binding protein-1;PDBTitle: the dna binding domain of rtbp1
62 c3rceA_
not modelled
10.3
26
PDB header: transferase/peptideChain: A: PDB Molecule: oligosaccharide transferase to n-glycosylate proteins;PDBTitle: bacterial oligosaccharyltransferase pglb
63 c2qzjC_
not modelled
10.2
18
PDB header: transcriptionChain: C: PDB Molecule: two-component response regulator;PDBTitle: crystal structure of a two-component response regulator from2 clostridium difficile
64 d2a9pa1
not modelled
10.2
23
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related65 c2rjnA_
not modelled
9.8
22
PDB header: hydrolaseChain: A: PDB Molecule: response regulator receiver:metal-dependentPDBTitle: crystal structure of an uncharacterized protein q2bku2 from2 neptuniibacter caesariensis
66 c2jrzA_
not modelled
9.7
18
PDB header: oxidoreductaseChain: A: PDB Molecule: histone demethylase jarid1c;PDBTitle: solution structure of the bright/arid domain from the human2 jarid1c protein.
67 c2ji6B_
not modelled
9.7
21
PDB header: lyaseChain: B: PDB Molecule: oxalyl-coa decarboxylase;PDBTitle: x-ray structure of oxalyl-coa decarboxylase in complex with2 3-deaza-thdp and oxalyl-coa
68 d1dbwa_
not modelled
9.5
20
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related69 c2hqoA_
not modelled
9.5
7
PDB header: signaling proteinChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: structure of a atypical orphan response regulator protein revealed a2 new phosphorylation-independent regulatory mechanism
70 c3j08A_
not modelled
9.5
19
PDB header: hydrolase, metal transportChain: A: PDB Molecule: copper-exporting p-type atpase a;PDBTitle: high resolution helical reconstruction of the bacterial p-type atpase2 copper transporter copa
71 d1q3ma_
not modelled
9.3
29
Fold: GLA-domainSuperfamily: GLA-domainFamily: GLA-domain72 c3lwfD_
not modelled
9.2
13
PDB header: transcription regulatorChain: D: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator (np_470886.1)2 from listeria innocua at 2.06 a resolution
73 c3gl9B_
not modelled
9.2
24
PDB header: signaling proteinChain: B: PDB Molecule: response regulator;PDBTitle: the structure of a histidine kinase-response regulator2 complex sheds light into two-component signaling and3 reveals a novel cis autophosphorylation mechanism
74 c2bpbB_
not modelled
9.1
23
PDB header: oxidoreductaseChain: B: PDB Molecule: sulfite\:cytochrome c oxidoreductase subunit b;PDBTitle: sulfite dehydrogenase from starkeya novella
75 c2jxjA_
not modelled
9.1
15
PDB header: oxidoreductaseChain: A: PDB Molecule: histone demethylase jarid1a;PDBTitle: nmr structure of the arid domain from the histone h3k42 demethylase rbp2
76 c3r0jA_
not modelled
8.8
22
PDB header: dna binding proteinChain: A: PDB Molecule: possible two component system response transcriptionalPDBTitle: structure of phop from mycobacterium tuberculosis
77 d1w36d1
not modelled
8.8
33
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain78 c3f6cB_
not modelled
8.7
13
PDB header: dna binding proteinChain: B: PDB Molecule: positive transcription regulator evga;PDBTitle: crystal structure of n-terminal domain of positive transcription2 regulator evga from escherichia coli
79 c3b2nA_
not modelled
8.7
9
PDB header: transcriptionChain: A: PDB Molecule: uncharacterized protein q99uf4;PDBTitle: crystal structure of dna-binding response regulator, luxr family, from2 staphylococcus aureus
80 c3hdgE_
not modelled
8.6
11
PDB header: structural genomics, unknown functionChain: E: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the n-terminal domain of an2 uncharacterized protein (ws1339) from wolinella3 succinogenes
81 c2panF_
not modelled
8.6
21
PDB header: lyaseChain: F: PDB Molecule: glyoxylate carboligase;PDBTitle: crystal structure of e. coli glyoxylate carboligase
82 c1xmeB_
not modelled
8.5
6
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c oxidase polypeptide ii;PDBTitle: structure of recombinant cytochrome ba3 oxidase from thermus2 thermophilus
83 c1k8vA_
not modelled
8.5
43
PDB header: unknown functionChain: A: PDB Molecule: neuropeptide f;PDBTitle: the nmr-derived conformation of neuropeptide f from2 moniezia expansa
84 c3eodA_
not modelled
8.4
20
PDB header: signaling proteinChain: A: PDB Molecule: protein hnr;PDBTitle: crystal structure of n-terminal domain of e. coli rssb
85 d1heya_
not modelled
8.2
11
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related86 c3hdlA_
not modelled
8.2
19
PDB header: oxidoreductaseChain: A: PDB Molecule: royal palm tree peroxidase;PDBTitle: crystal structure of highly glycosylated peroxidase from royal palm2 tree
87 c2nxwB_
not modelled
8.2
31
PDB header: lyaseChain: B: PDB Molecule: phenyl-3-pyruvate decarboxylase;PDBTitle: crystal structure of phenylpyruvate decarboxylase of azospirillum2 brasilense
88 d1xg7a_
not modelled
8.1
7
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: AgoG-like89 d1qh8a_
not modelled
8.1
11
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein90 d1pa2a_
not modelled
8.0
26
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: CCP-like91 d1o6ea_
not modelled
8.0
19
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin92 c1hr6C_
not modelled
7.9
11
PDB header: hydrolaseChain: C: PDB Molecule: mitochondrial processing peptidase alpha subunit;PDBTitle: yeast mitochondrial processing peptidase
93 c3cg0A_
not modelled
7.6
11
PDB header: lyaseChain: A: PDB Molecule: response regulator receiver modulated diguanylate cyclasePDBTitle: crystal structure of signal receiver domain of modulated diguanylate2 cyclase from desulfovibrio desulfuricans g20, an example of alternate3 folding
94 c3mmnA_
not modelled
7.6
14
PDB header: transferaseChain: A: PDB Molecule: histidine kinase homolog;PDBTitle: crystal structure of the receiver domain of the histidine kinase cki12 from arabidopsis thaliana complexed with mg2+
95 d2djia1
not modelled
7.5
26
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain96 c2pgnA_
not modelled
7.5
31
PDB header: hydrolaseChain: A: PDB Molecule: cyclohexane-1,2-dione hydrolase (cdh);PDBTitle: the crystal structure of fad and thdp-dependent cyclohexane-1,2-dione2 hydrolase in complex with cyclohexane-1,2-dione
97 d1qkka_
not modelled
7.3
16
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related98 c1fp2A_
not modelled
7.3
15
PDB header: transferaseChain: A: PDB Molecule: isoflavone o-methyltransferase;PDBTitle: crystal structure analysis of isoflavone o-methyltransferase
99 d2ev0a1
not modelled
7.2
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Iron-dependent repressor protein